Which genes are different?
Read the Full Genome of C. rudii
Please download from NCBI the file AP009180.fna,
containing the full genome of Carsonella rudii. The genome
FASTA file contains one sequence for each chromosome. Read it into a
list called chromosomes session and show how many
chromosomes you got.
[1] 1# write hereTake the genome from the chromosomes
Remember that read.fasta will always give you a list.
For example, a list of all genes, a list of all proteins, a list of all
DNA reads from the sequencing machine, or a list of all chromosomes. In
most cases you want to use all elements of the list.
Since in this case we have only one chromosome, it is practical to
assign it to a vector called genome.
# write hereNow you can show the length of this vector.
length(genome)[1] 159662Genome GC content
Now you can calculate the genome GC content (i.e. the mean
of is_G_or_C event) and assign it to
gc_genome. You can also calculate the variance of
is_G_or_C and assign it to var_gc_genome. You
need to use again the same is_G_or_C function that you used
in the main part of Homework 11.
# write hereThey values should be these:
gc_genome[1] 0.1656437var_gc_genome[1] 0.1382067This allow you to calculate how much variation of GC content is expected if the genes are random. We are lucky this time: you know the population variance, so you can predict the standard error of the GC[^stderr] of each gene as the standard deviation of the population divided by the square root of the gene length.
Genes
Now you must download the file c_rudii.fna.txt
and save it in your computer. It has to be stored in your computer to be
fast and efficient.
Using the library seqinr, please write the code to read
the FASTA file and load all genes in a list called genes.
Then calculate the length of each gene and store them in a vector called
gene_length. You also need the GC content of each gene,
that you should store in the vector gc_genesYou can use the function GC() from the
seqinr library, or use
mean(is_G_or_C(gene)).
# write hereTest your results with this code:
length(genes)[1] 182hist(gene_length, col="grey")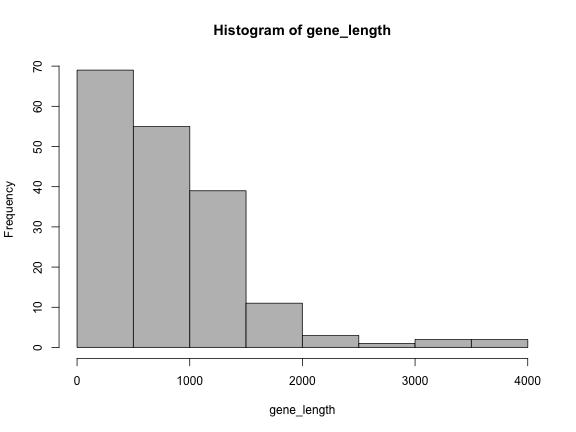
hist(gc_genes, col="grey")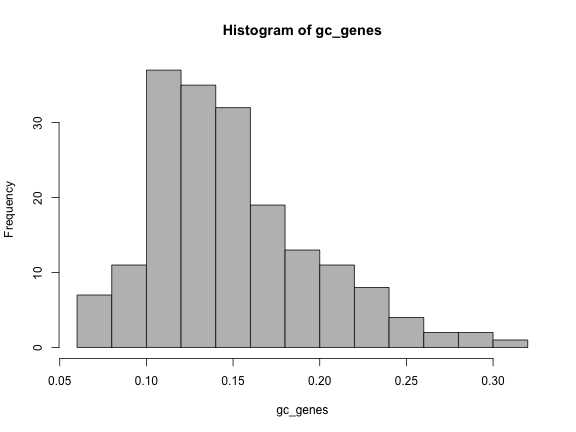
Confidence intervals
Since the GC content is an average of many things, all with the same
variance, then the genes GC content will follow a Normal distribution.
Therefore you can calculate the confidence interval width using the
qnorm() functionRemember that this time you know the population
variance
.
Please calculate the vectors top and
bottom, containing the limits of the confidence interval
for each gene. Considere a confidence level of \((1-\alpha)=99.9\%.\)
Then you can see them in a plot like this:
plot(top, type="l", ylim=c(0, 0.3))
abline(h=gc_genome)
lines(bottom)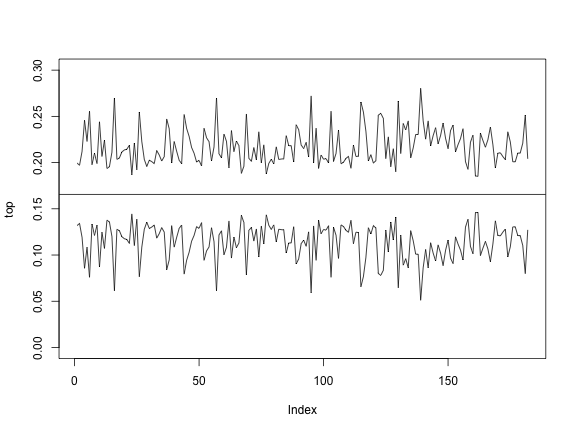
Atypical genes
These confidence intervals tells us how much variation can we expect
in the GC content of a gene if it was a random sample of the genome. Any
gene with GC content below bottom or over top
are atypical. They have a very low probability of being just a
random sample. Therefore, we often assume they are different due to a
biological reason.
Please show which genes have GC content below bottom
BAF35036.1 BAF35037.1 BAF35045.1 BAF35052.1 BAF35058.1 BAF35065.1
5 6 14 21 27 34
BAF35066.1 BAF35067.1 BAF35071.1 BAF35073.1 BAF35078.1 BAF35079.1
35 36 40 42 47 48
BAF35081.1 BAF35085.1 BAF35087.1 BAF35090.1 BAF35101.1 BAF35102.1
50 54 56 59 70 71
BAF35106.1 BAF35117.1 BAF35119.1 BAF35124.1 BAF35127.1 BAF35128.1
75 86 88 93 96 97
BAF35132.1 BAF35133.1 BAF35140.1 BAF35142.1 BAF35145.1 BAF35154.1
101 102 109 111 114 123
BAF35157.1 BAF35160.1 BAF35196.1 BAF35202.1 BAF35204.1 BAF35205.1
126 129 165 171 173 174
BAF35207.1 BAF35211.1 BAF35213.1
176 180 182 # write hereand which genes have GC content over top
BAF35038.1 BAF35040.1 BAF35060.1 BAF35082.1 BAF35093.1 BAF35103.1
7 9 29 51 62 72
BAF35108.1 BAF35110.1 BAF35111.1 BAF35113.1 BAF35135.1 BAF35139.1
77 79 80 82 104 108
BAF35156.1 BAF35168.1 BAF35184.1 BAF35188.1 BAF35189.1 BAF35191.1
125 137 153 157 158 160
BAF35192.1 BAF35212.1
161 181 # write here