Class 17: Normalization in Arrays
Systems Biology
Andrés Aravena, PhD
December 21, 2021
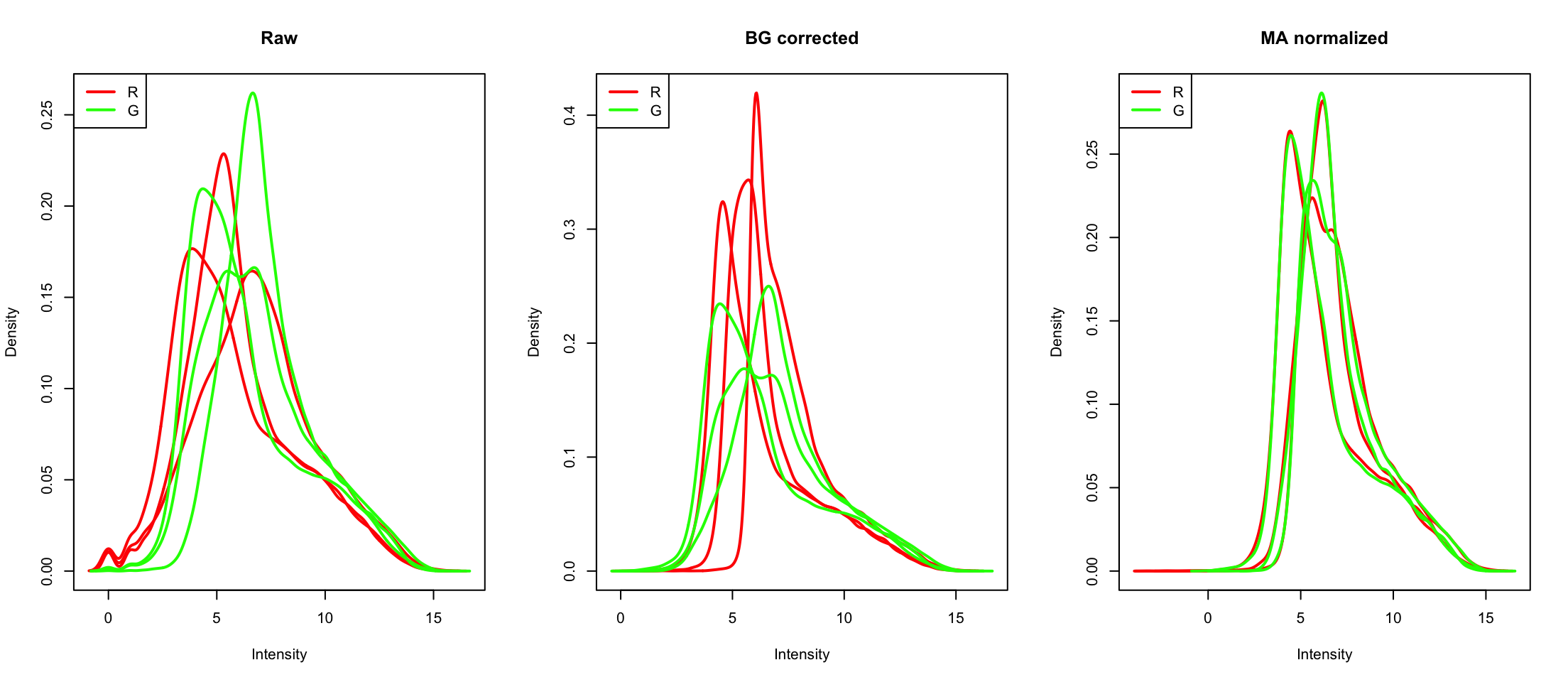
Normalization
- Background correction
- Remove technical noise
- Within arrays
- Guarantee that observed differential expression is real
- “Makes average equal to zero”
- Between arrays
- Enables comparison between conditions
- “Makes variances equal”
Within arrays
- Uses logarithm of war expression
- Remove background signal
- Assumes that some genes are not differentially expressed
- Either housekeeping genes or most of the genes
- they are forced to have zero differential expression
Let’s read some data
library(limma)
targets <- readTargets("targets.txt")
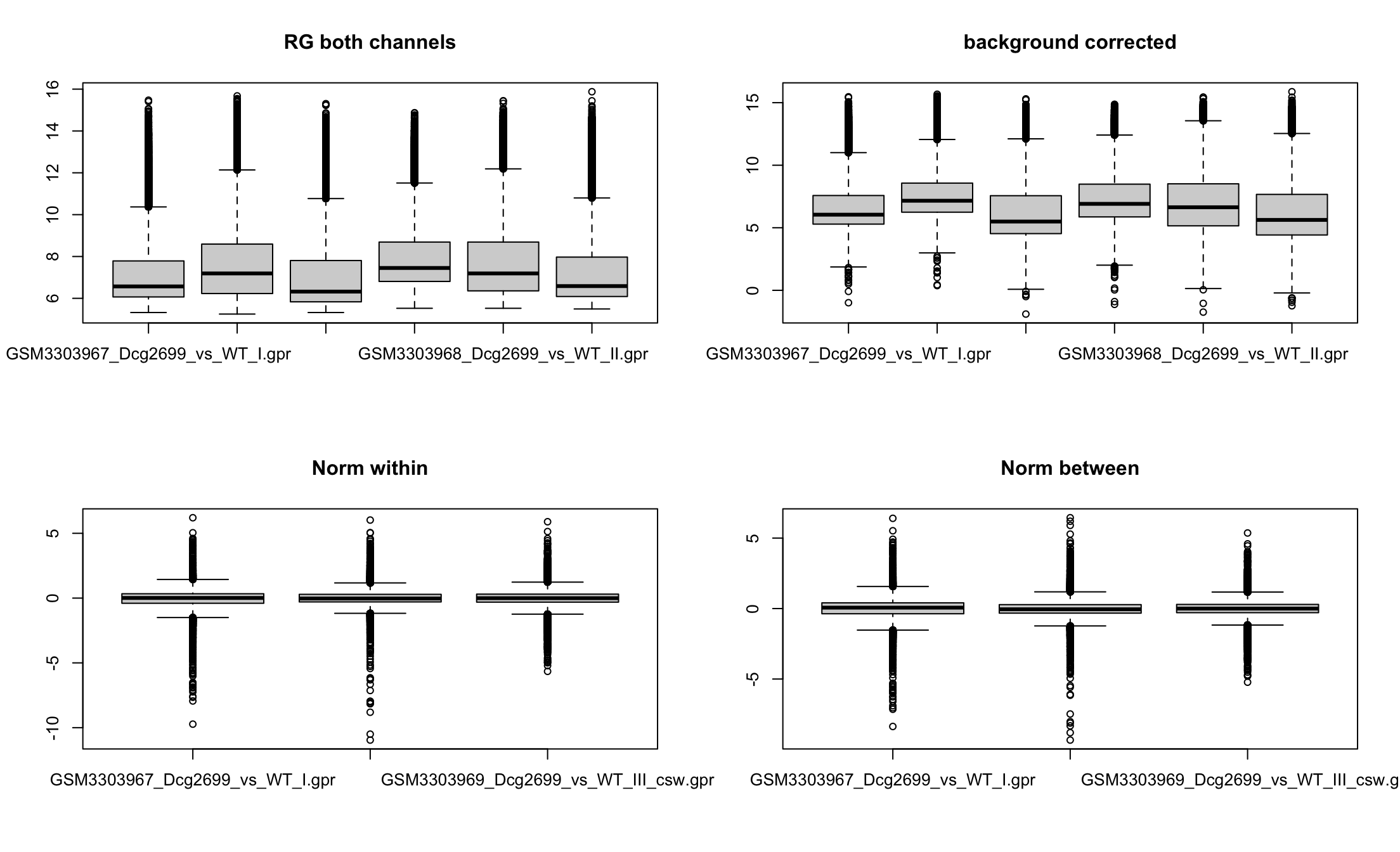
RG <- read.maimages(targets$Filename, source = "genepix")Read GSM3303967_Dcg2699_vs_WT_I.gpr.gz
Read GSM3303968_Dcg2699_vs_WT_II.gpr.gz
Read GSM3303969_Dcg2699_vs_WT_III_csw.gpr.gz Array 1 corrected
Array 2 corrected
Array 3 corrected
Array 1 corrected
Array 2 corrected
Array 3 correctedBox plots
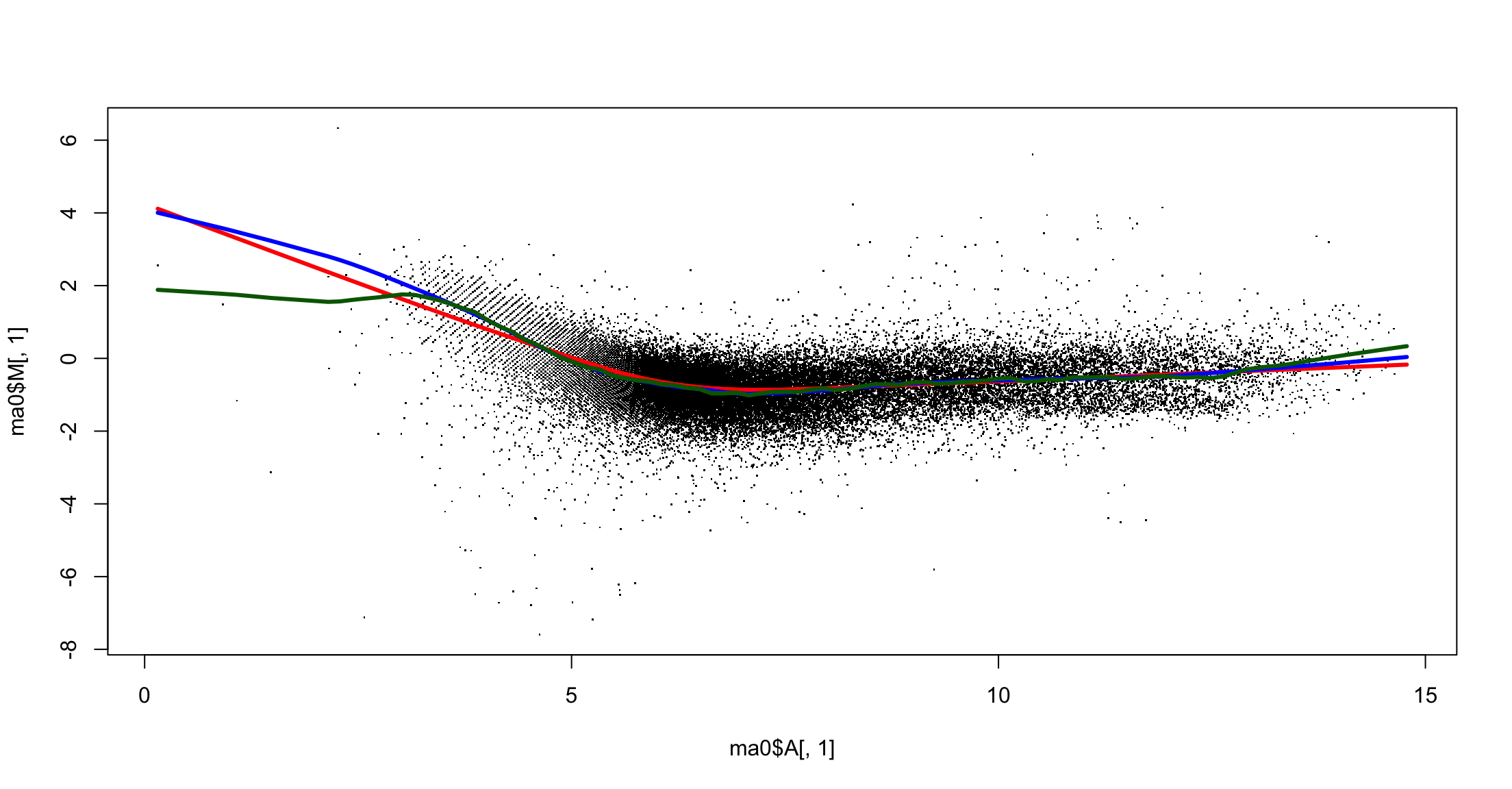
Lowess: within array normalization
Pending
NormExp model for background normalization
Combining probes for a single gene
Clustering
Heathmap
Single color arrays
Affymetrix
Massive arrays
- Hundreds of thousands of spots
In situ synthesis
Each oligo has a negative control
Negative control
Oligos are short. Let’s say 25 bp
There may be several oligos for each gene
Positive match oligos are called \(PM\)
For each \(PM\) there is a negative control called \(MM\)
\(MM\) oligos have a mismatching basepair in the center bp
- Position 13 if oligos arr 25 bp long
Normalization
The \(MM\) probe will not hybridize with the gene, but will hybridize with random fragments
The same random fragments will hybridize with the \(PM\)
The difference \(PM-MM\) is the signal of the gene