Class 16: Analyzing Two-color Microarrays
Systems Biology
Andrés Aravena, PhD
December 17, 2021
Following the limma protocol
For today’s class we will follow the protocol for two-colors microarrays
- Define the experiment
- Read the hybridization data
- Normalize each slide internally
- Normalize between arrays
- Fit a linear model
- Evaluate statistical significance
Targets
It is recommended to start with a description of the experiments
We write a tab-separated file, indicating at least
- Filename
- Cy3
- Cy5
Filename Cy3 Cy5
1 GSM3303967_Dcg2699_vs_WT_I.gpr.gz mutant wt
2 GSM3303968_Dcg2699_vs_WT_II.gpr.gz mutant wt
3 GSM3303969_Dcg2699_vs_WT_III_csw.gpr.gz wt mutantReading the targets file
| Filename | Cy3 | Cy5 |
|---|---|---|
| GSM3303967_Dcg2699_vs_WT_I.gpr.gz | mutant | wt |
| GSM3303968_Dcg2699_vs_WT_II.gpr.gz | mutant | wt |
| GSM3303969_Dcg2699_vs_WT_III_csw.gpr.gz | wt | mutant |
This example was taken from GSE117566 in NCBI GEO
Reading the arrays
Read GSM3303967_Dcg2699_vs_WT_I.gpr.gz
Read GSM3303968_Dcg2699_vs_WT_II.gpr.gz
Read GSM3303969_Dcg2699_vs_WT_III_csw.gpr.gz [1] "RGList"
attr(,"package")
[1] "limma"[1] "R" "G" "Rb" "Gb" "targets" "genes" "source"
[8] "printer"[1] 45220 3Genes
Block Row Column ID Name
1 1 1 1 GE_BrightCorner GE_BrightCorner
2 1 1 2 GE_BrightCorner GE_BrightCorner
3 1 1 3 DarkCorner DarkCorner
4 1 1 4 DarkCorner DarkCorner
5 1 1 5 DarkCorner DarkCorner
6 1 1 6 DarkCorner DarkCorner
7 1 1 7 DarkCorner DarkCorner
8 1 1 8 DarkCorner DarkCorner
9 1 1 9 DarkCorner DarkCorner
10 1 1 10 DarkCorner DarkCorner
11 1 1 11 DarkCorner DarkCorner
12 1 1 12 DarkCorner DarkCorner
13 1 1 13 DarkCorner DarkCorner
14 1 1 14 DarkCorner DarkCorner
15 1 1 15 DarkCorner DarkCornerComparing R and G channels
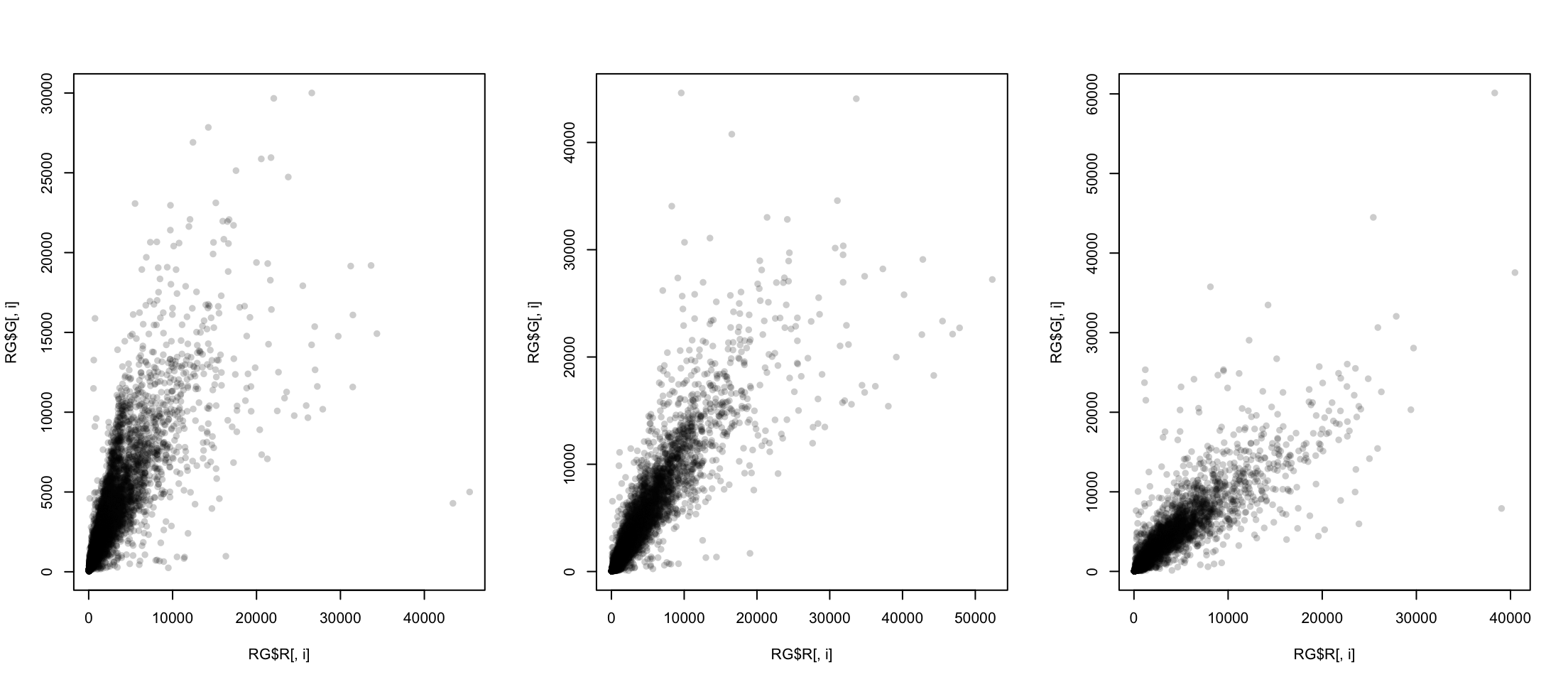
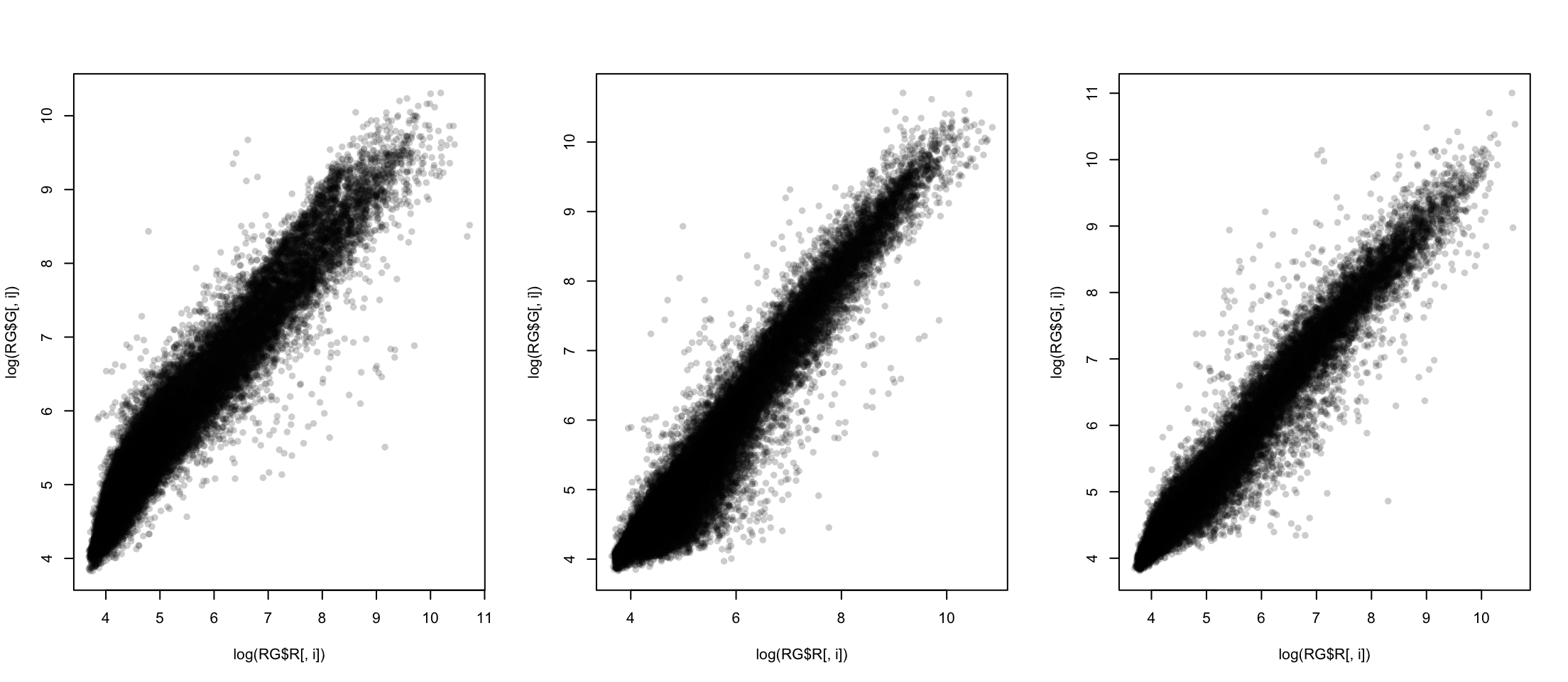
Using Logarithms
Looking at the slides
Using log
for(i in 1:3) imageplot(log(RG$R[,i]), RG$printer)
for(i in 1:3) imageplot(log(RG$G[,i]), RG$printer)
Background
for(i in 1:3) imageplot(log(RG$Rb[,i]), RG$printer)
for(i in 1:3) imageplot(log(RG$Gb[,i]), RG$printer)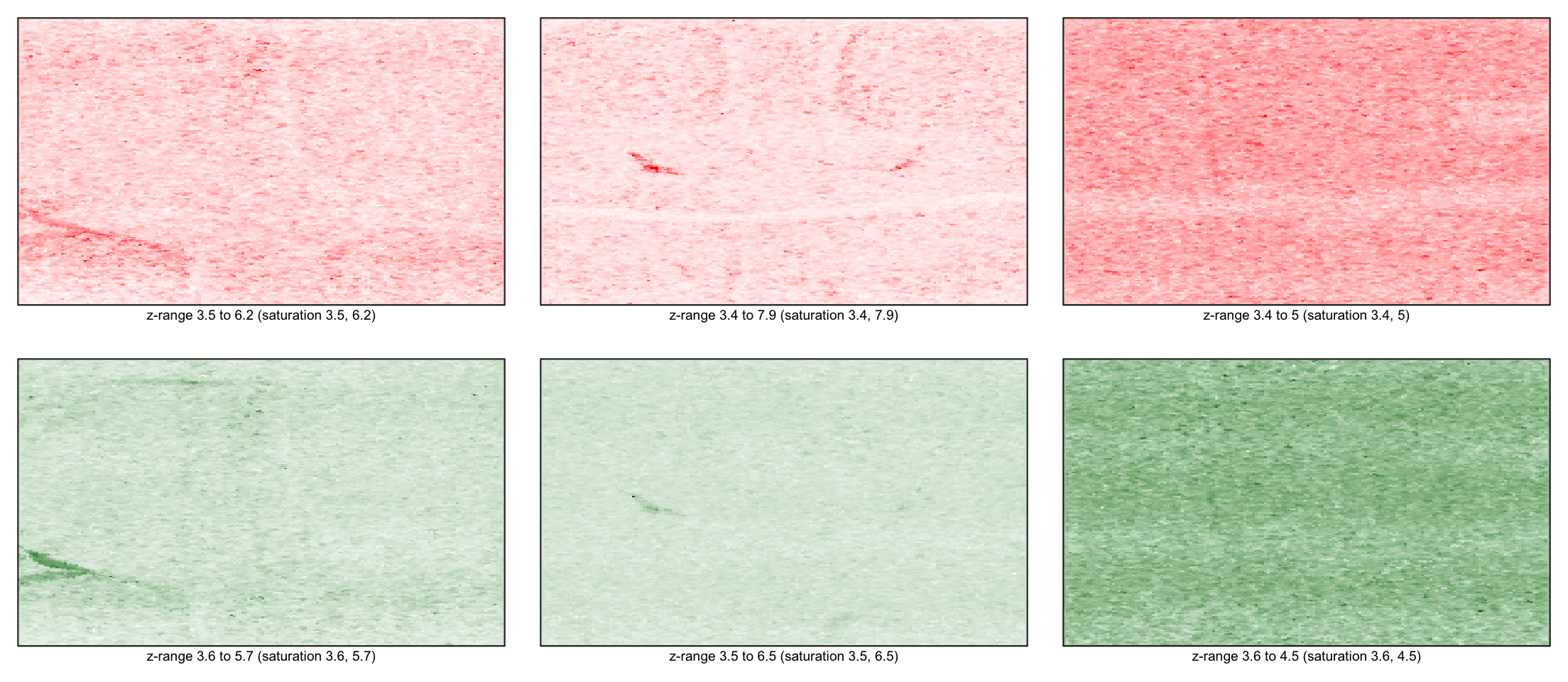
Background correction
The easiest way to normalize is to subtract the background from the signal \[R_\text{corrected} = R - Rb\]
[1] "RGList"
attr(,"package")
[1] "limma"[1] "R" "G" "targets" "genes" "source" "printer"Histogram
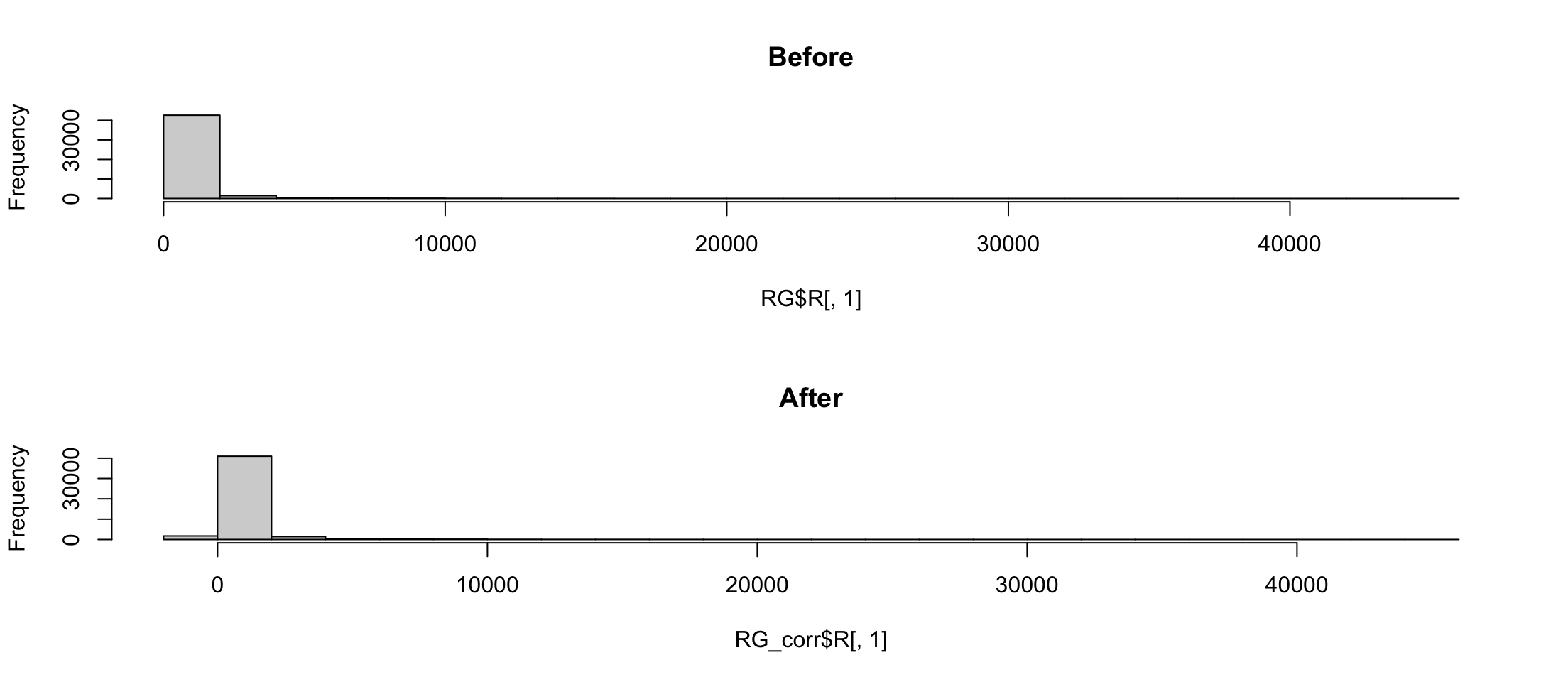
There are negative numbers. Not good for logarithms
Histogram of log
Warning in log(RG_corr$R[, 1]): NaNs produced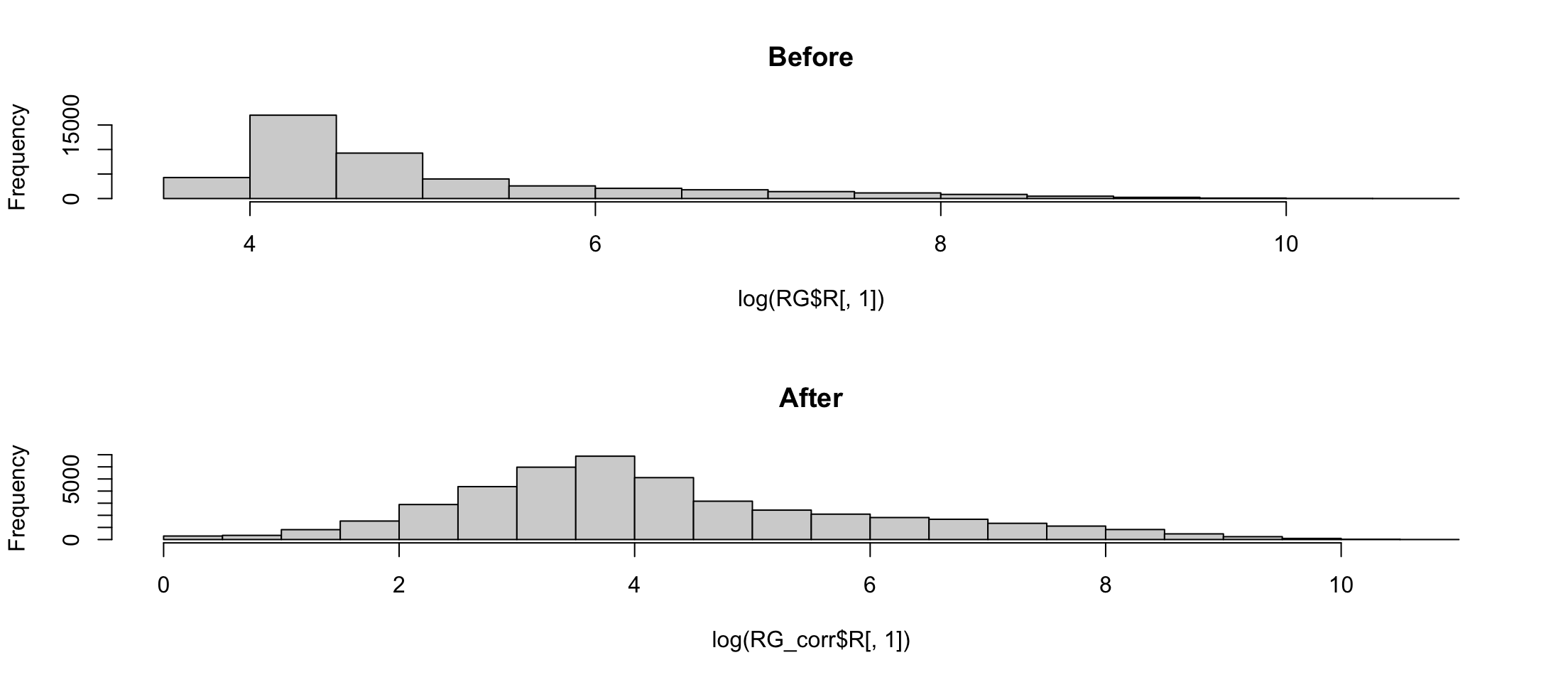
Avoiding negative values
A better correction requires a better model
Array 1 corrected
Array 2 corrected
Array 3 corrected
Array 1 corrected
Array 2 corrected
Array 3 correctedNormexp uses a model of background
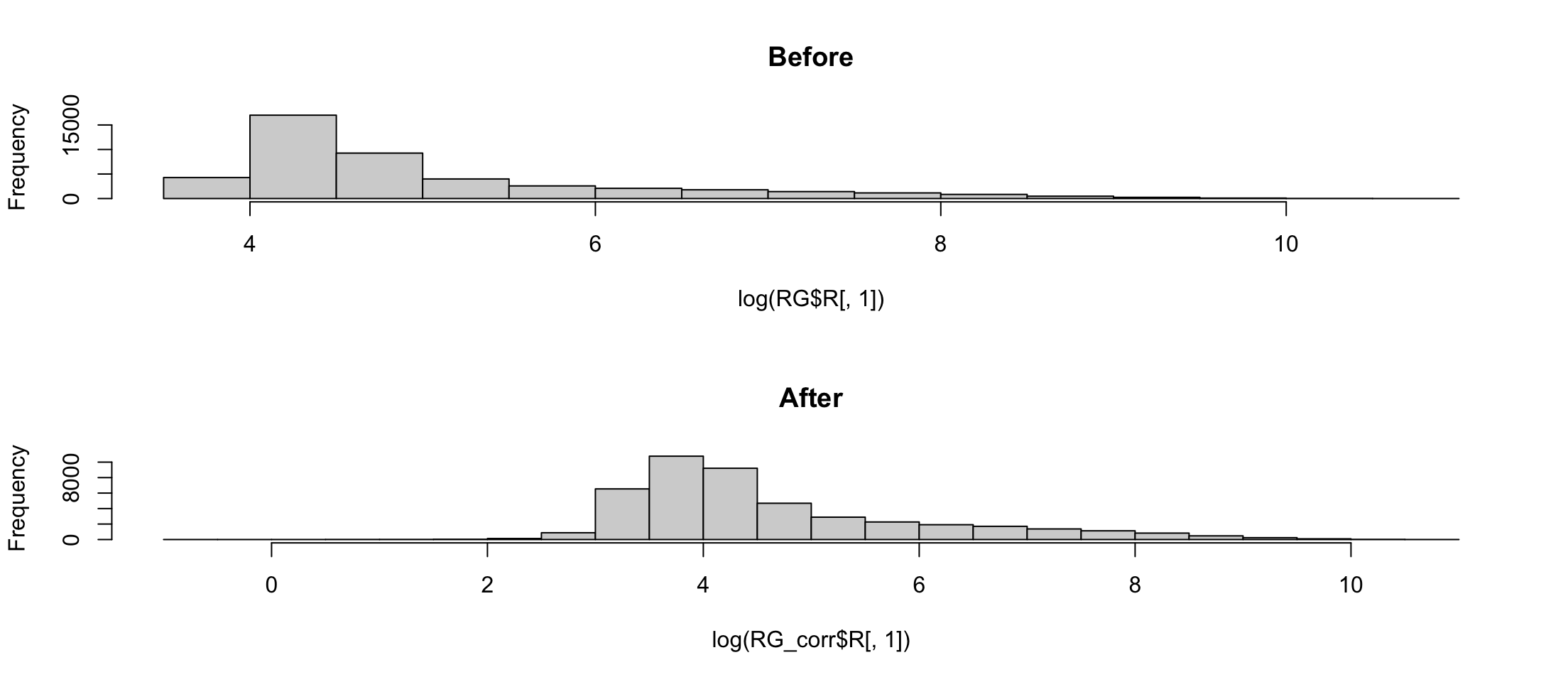
Background corrected images
for(i in 1:3) imageplot(log(RG$R[,i]), RG$printer)
for(i in 1:3) imageplot(log(RG$G[,i]), RG$printer)
Comparing corrected
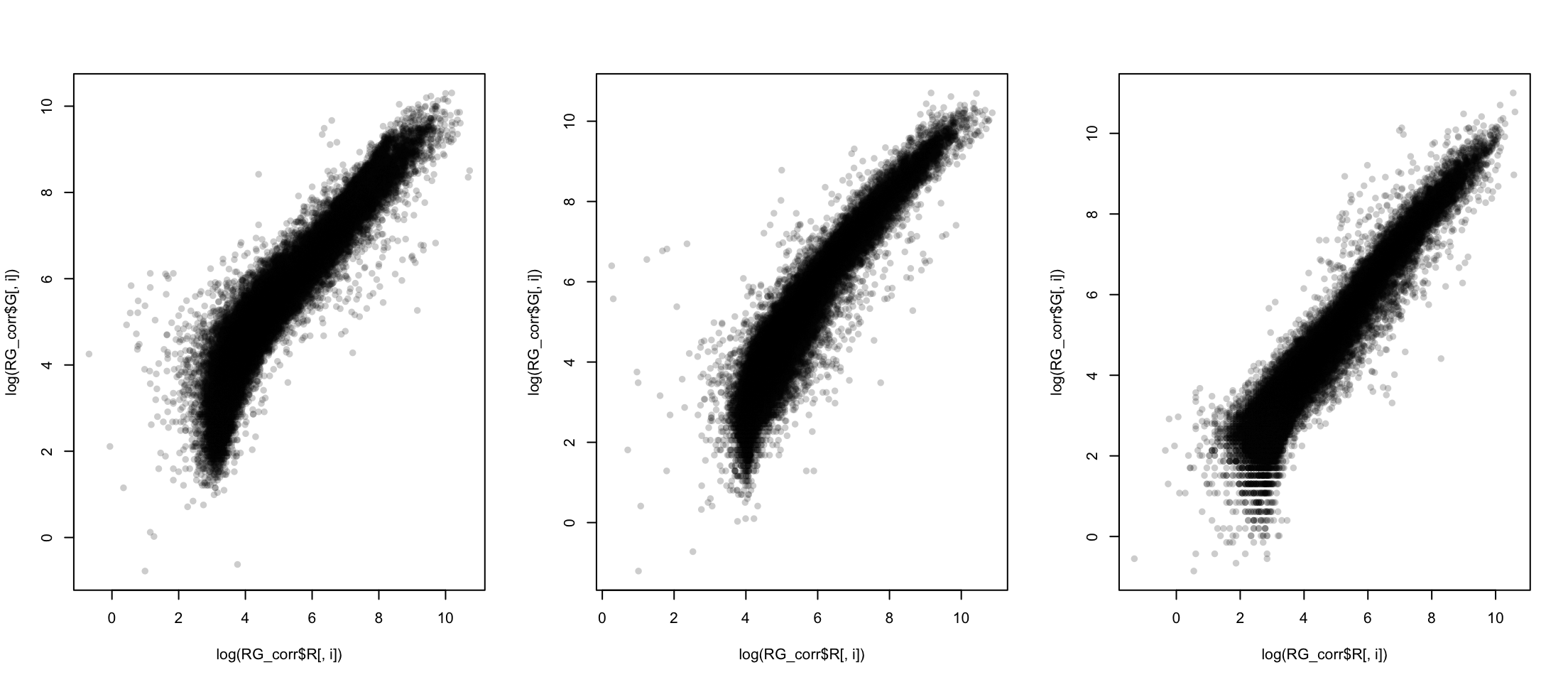
Changing perspective
We care about the difference of expression \[M = \frac{\log(R)-\log(G)}{2}\]
For symmetry, we also keep the average expression \[ A = \frac{\log(R)+\log(G)}{2}\]
MA plot
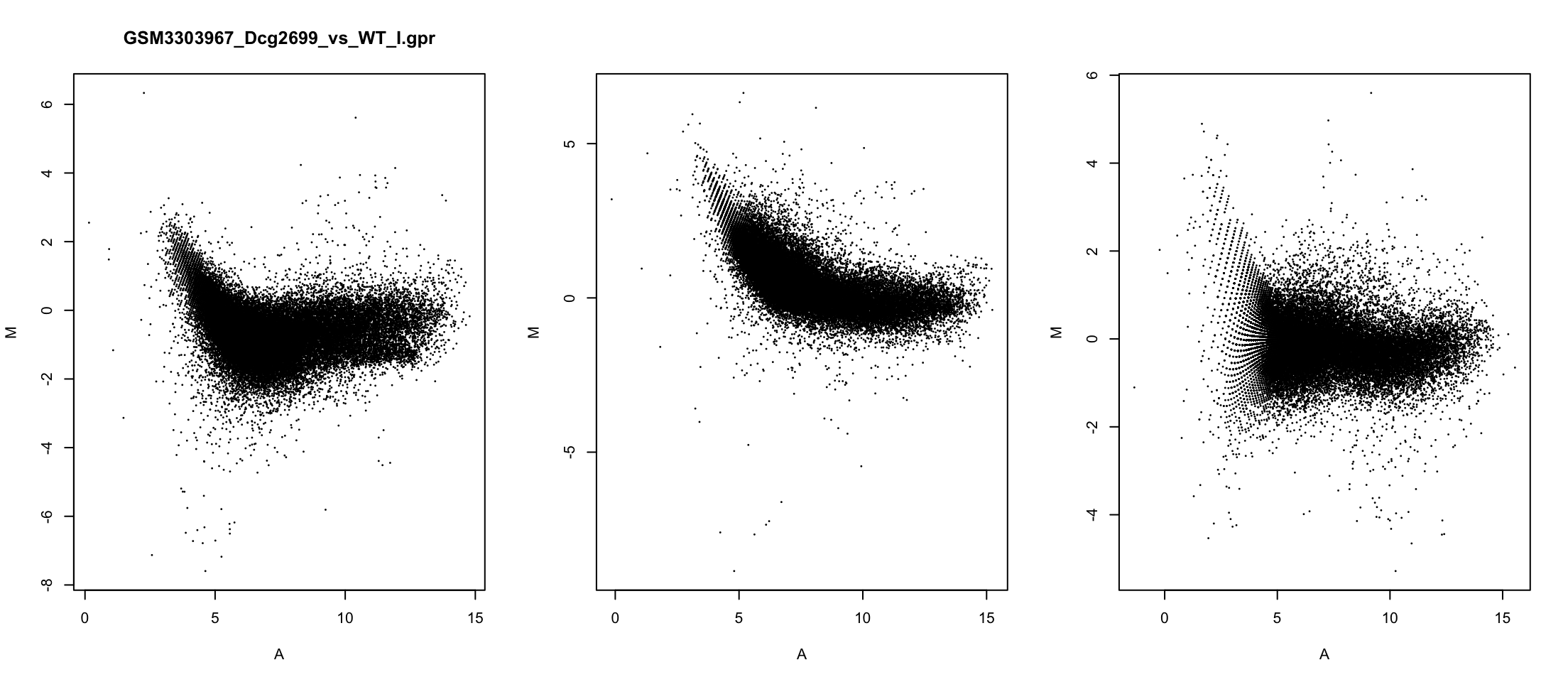
Correcting banana shape
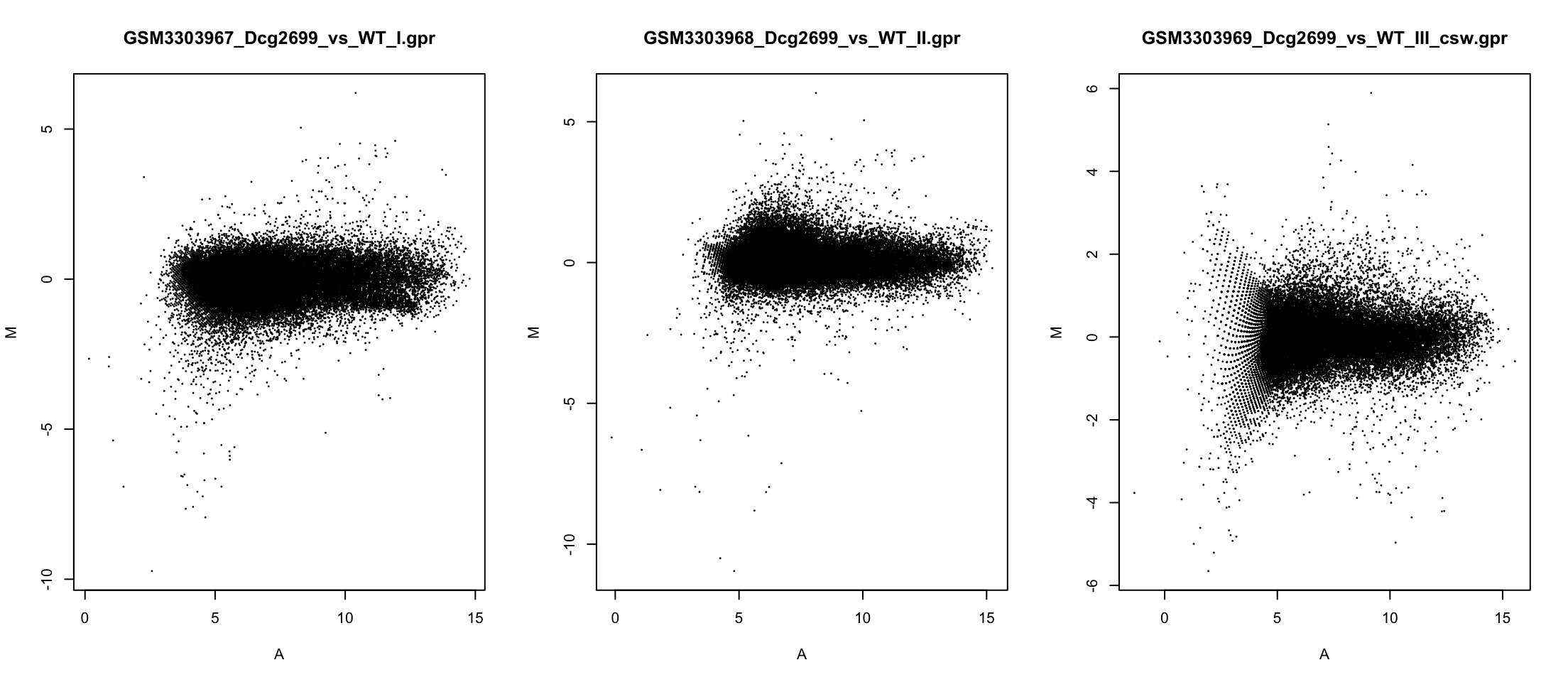
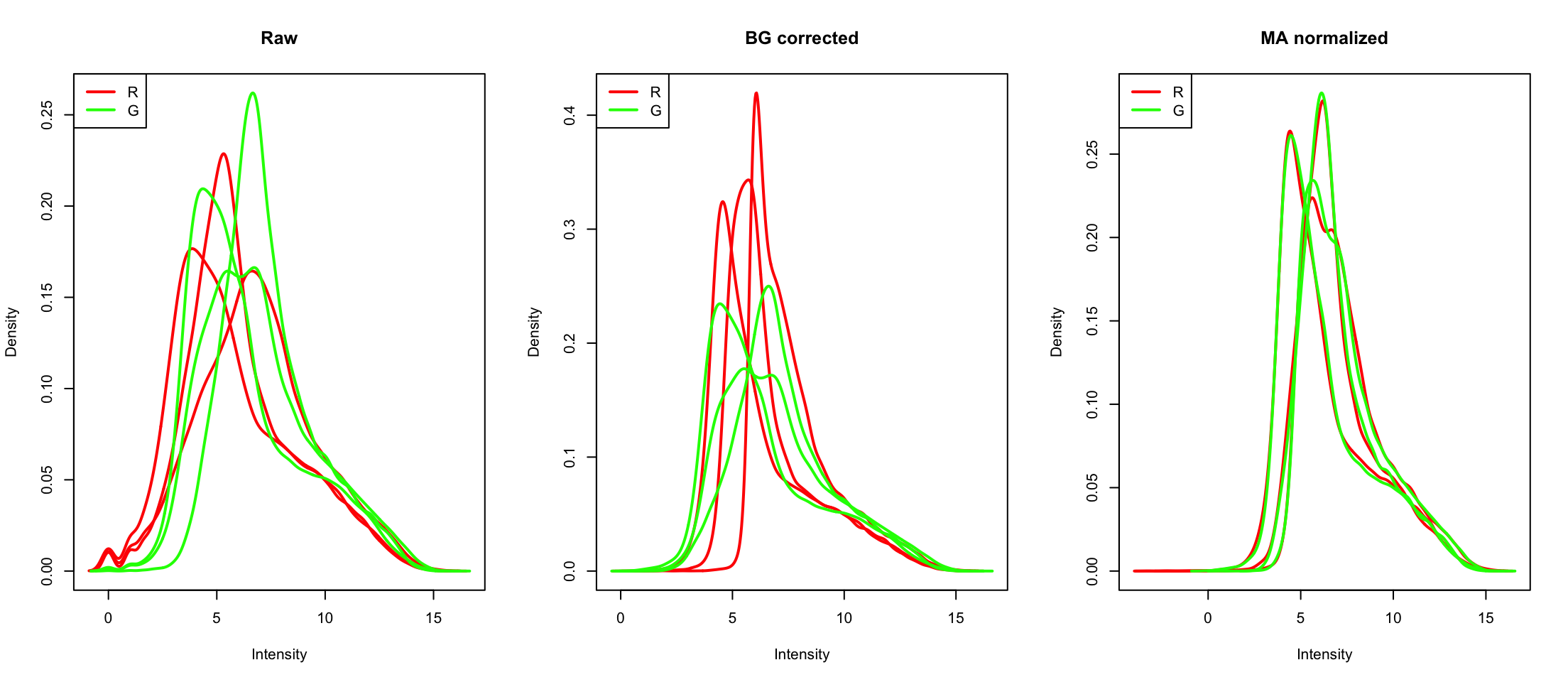
Comparing densities
Think of these as softened histograms
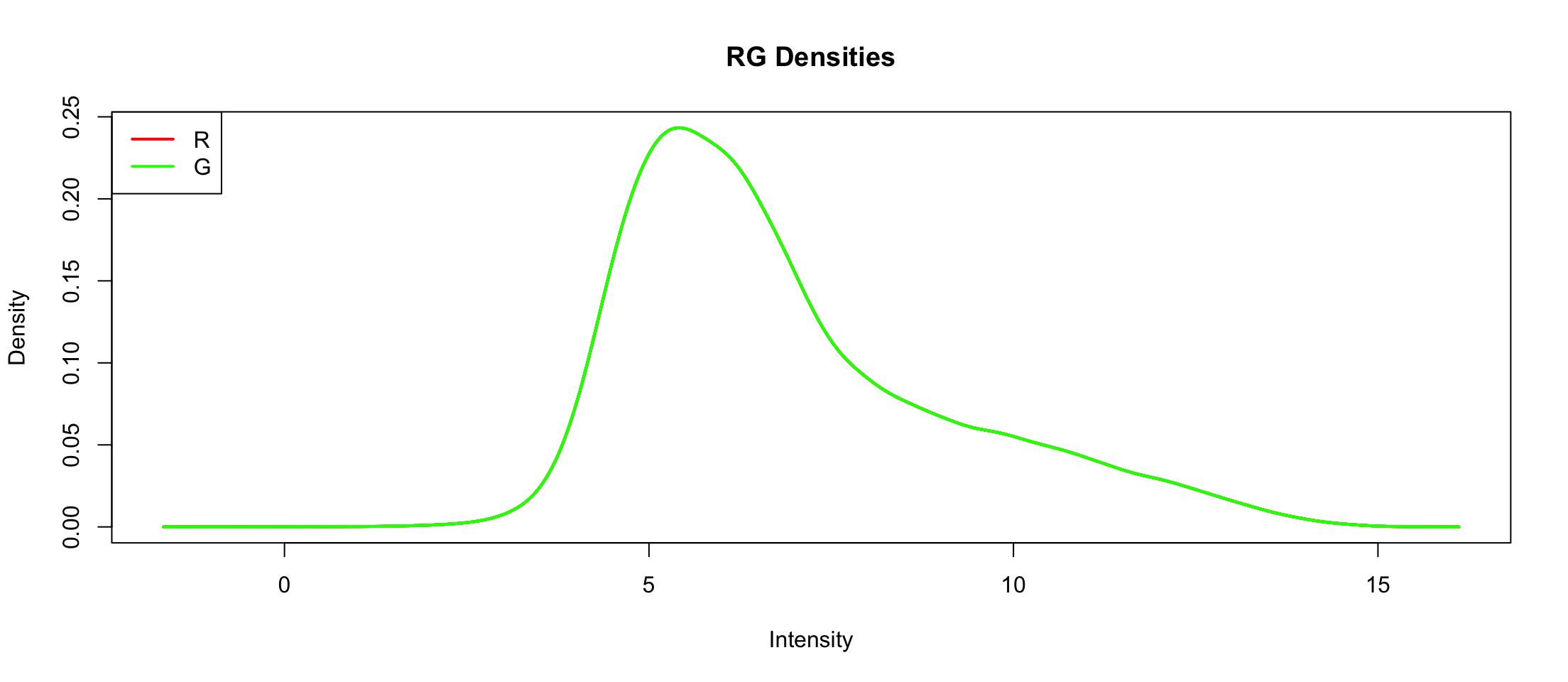
Between array normalization
Let’s drop control spots
gal <- readGAL("GPL16989_Genepix_GAL_4plex_Design_041487.gal.gz")
# num_replicas <- table(MA$genes$ID)
# controls <- names(num_replicas)[num_replicas>=10]
MA <- MA[gal$ControlType == "false", ]
MA <- MA[MA$genes$ID != "EMPTY", ]
MA.q <- normalizeBetweenArrays(MA, method = "quantile")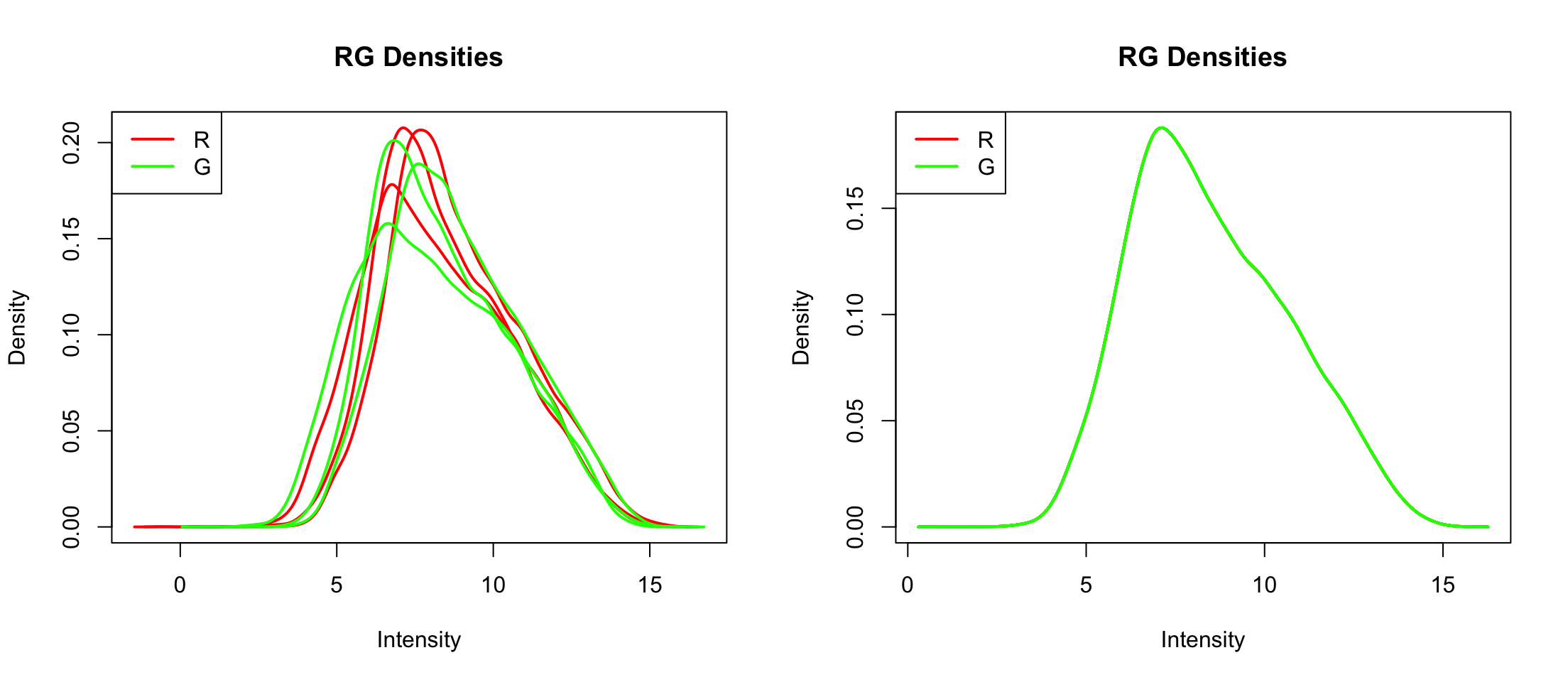
Now we are ready to analyze
First, simplify gene description
There are many columns that we do not need anymore
Let’s drop them
This will simplify the presentation of results later
We can build a model matrix from targets
Found unique target names:
mutant wt mutant
[1,] -1
[2,] -1
[3,] 1Now we fit it
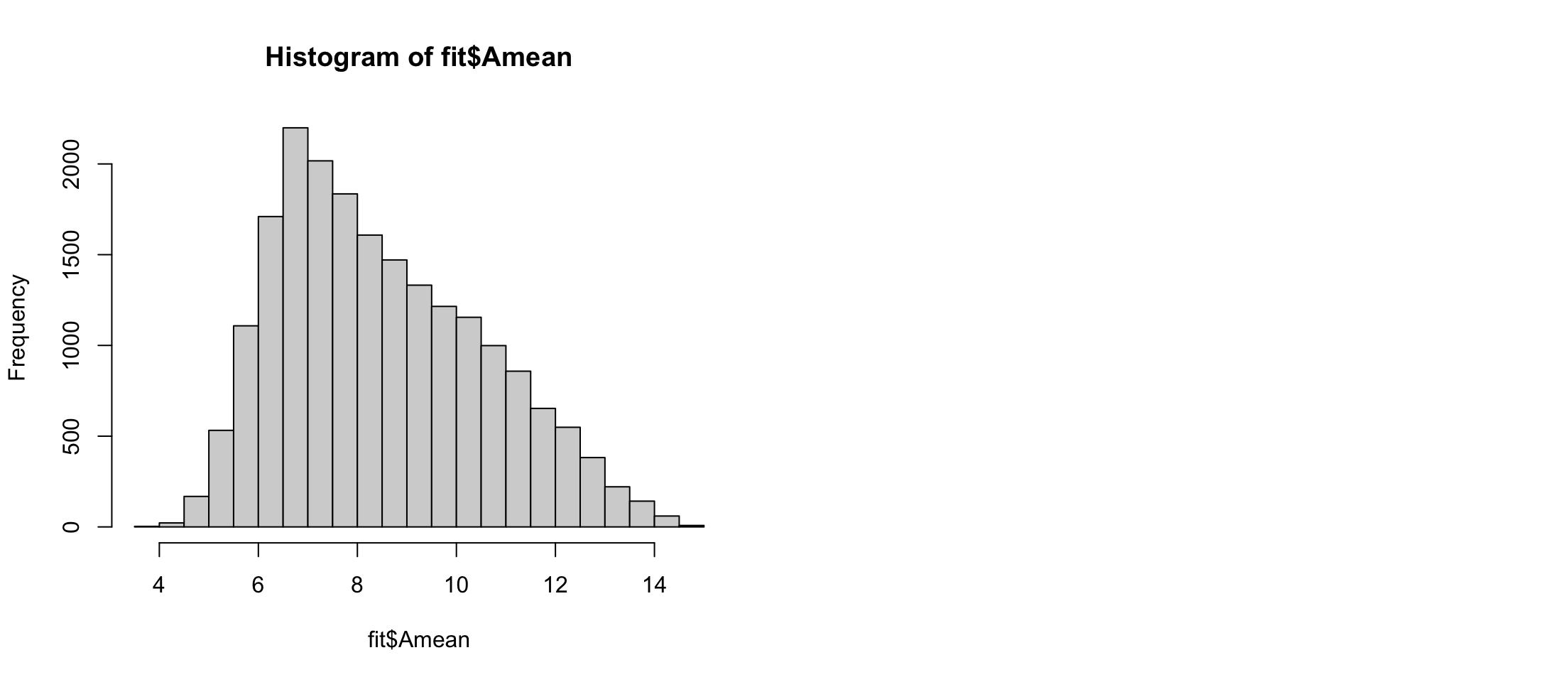
Filter out the un-expressed spots
Name logFC AveExpr t P.Value adj.P.Val
1039 cg2071 5.428600 9.440261 26.03824 1.025257e-10 2.056051e-06
24758 TOCGc27704421 -5.412790 10.218971 -20.27835 1.277049e-09 1.109858e-05
42170 cg1299 -3.877140 10.355572 -18.36246 3.450906e-09 1.109858e-05
25823 cg1883 -3.955607 11.980194 -18.31029 3.550412e-09 1.109858e-05
18653 cg1881 -4.016262 11.966937 -18.00908 4.189938e-09 1.109858e-05
33607 cg1881 -3.851484 10.706278 -17.97686 4.265475e-09 1.109858e-05
7177 cg1883 -4.016393 11.083149 -17.57707 5.338004e-09 1.109858e-05
28545 cg1299 -3.878424 10.628074 -17.57305 5.350168e-09 1.109858e-05
37305 cg1884 -3.775995 10.826130 -17.54943 5.422361e-09 1.109858e-05
117 cg1881 -4.023286 12.212845 -17.48055 5.639087e-09 1.109858e-05
B
1039 13.50041
24758 11.84673
42170 11.10935
25823 11.08761
18653 10.96019
33607 10.94637
7177 10.77173
28545 10.76995
37305 10.75944
117 10.72868