Class 15: Practice with Interaction Graphs
Systems Biology
Andrés Aravena, PhD
December 14, 2021
An example of microarray analysis
This is Atakan’s thesis work
We start with a description of the experiments
It is an Excel table, which we export as tab-separated file and read it in R
[1] 81 10We get a data frame with 81 rows and 10 columns
Let’s see the content
individual organism diet age..week. organism.part material.type label
1 T95 Mus musculus AL 49/50 Brain organism part biotin
2 T96 Mus musculus AL 49/50 Brain organism part biotin
3 T207 Mus musculus AL 49/50 Brain organism part biotin
4 T106 Mus musculus CCR 49/50 Brain organism part biotin
5 T184 Mus musculus CCR 49/50 Brain organism part biotin
6 T185 Mus musculus CCR 49/50 Brain organism part biotin
array.design array.data.file
1 miRNA-4_0 Brain/AL_T95_Brain_w50.CEL
2 miRNA-4_0 Brain/AL_T96_Brain_w50.CEL
3 miRNA-4_0 Brain/AL_T207_Brain_w50.CEL
4 miRNA-4_0 Brain/CCR_T106_Brain_w50.CEL
5 miRNA-4_0 Brain/CCR_T184_brain_w50.CEL
6 miRNA-4_0 Brain/CCR_T185_Brain_w50.CEL
array.data.path
1 ~/CR-miRNA/Data/Brain/AL_T95_Brain_w50.CEL
2 ~/CR-miRNA/Data/Brain/AL_T96_Brain_w50.CEL
3 ~/CR-miRNA/Data/Brain/AL_T207_Brain_w50.CEL
4 ~/CR-miRNA/Data/Brain/CCR_T106_Brain_w50.CEL
5 ~/CR-miRNA/Data/Brain/CCR_T184_brain_w50.CEL
6 ~/CR-miRNA/Data/Brain/CCR_T185_Brain_w50.CELThey used two platforms
The microarray slides changed during the experiments
miRNA-4_0 miRNA-4_1
12 69 That will make the analysis more complicated
For today we will use only one platforms
How to read array data
There are several libraries for different platforms
We will see them later
For this data we use two libraries
The last library has the description of the microarray probes
Reading the data
Reading in : Brain/AL_T3_Brain_w80.ga.cel
Reading in : Brain/AL_T67_Brain_w80.ga.cel
Reading in : Brain/AL_T123_Brain_w80.ga.cel
Reading in : Brain/CCR_T33_Brain_w80.ga.cel
Reading in : Brain/CCR_T38_Brain_w80.ga.cel
Reading in : Brain/CCR_T125_Brain_w80.ga.cel
Reading in : Brain/ICRR_T137_Brain_w80.ga.cel
Reading in : Brain/ICRR_T70_Brain_w80.ga.cel
Reading in : Brain/ICRR_T22_Brain_w80.ga.cel
Reading in : Brain/ICRRF_T117_Brain_w80.ga.cel
Reading in : Brain/ICRRF_T116_Brain_w80.ga.cel
Reading in : Brain/ICRRF_T25_Brain_w80.ga.cel
Reading in : Blood/AL_T95_blood_w50.ga.cel
Reading in : Blood/AL_T96_blood_w50.ga.cel
Reading in : Blood/AL_T97_blood_w50.ga.cel
Reading in : Blood/CCR_T94_blood_w50.ga.cel
Reading in : Blood/CCR_T106_blood_w50.ga.cel
Reading in : Blood/CCR_T107_blood_w50.ga.cel
Reading in : Blood/ICRR_T79_blood_w50.ga.cel
Reading in : Blood/ICRR_T196_blood_w50.ga.cel
Reading in : Blood/ICRR_T197_blood_w50.ga.cel
Reading in : Blood/ICRRF_T76_blood_w50.ga.cel
Reading in : Blood/ICRRF_T111_blood_w50.ga.cel
Reading in : Blood/ICRRF_T223_blood_w50.ga.cel
Reading in : Blood/AL_T66_Blood_w80.ga.cel
Reading in : Blood/AL_T67_Blood_w80.ga.cel
Reading in : Blood/AL_T123_Blood_w80.ga.cel
Reading in : Blood/CCR_T38_Blood_w80.ga.cel
Reading in : Blood/CCR_T125_Blood_w80.ga.cel
Reading in : Blood/CCR_T126_Blood_w80.ga.cel
Reading in : Blood/ICRR_T70_Blood_w80.ga.cel
Reading in : Blood/ICRR_T137_Blood_w80.ga.cel
Reading in : Blood/ICRR_T138_Blood_w80.ga.cel
Reading in : Blood/ICRRF_T116_Blood_w80.ga.cel
Reading in : Blood/ICRRF_T117_Blood_w80.ga.cel
Reading in : Blood/ICRRF_T129_Blood_w80.ga.cel
Reading in : MFP/AL_T95_MFP_w50.ga.cel
Reading in : MFP/AL_T96_MFP_w50.ga.cel
Reading in : MFP/AL_T97_MFP_w50.ga.cel
Reading in : MFP/CCR_T94_MFP_w50.ga.cel
Reading in : MFP/CCR_T106_MFP_w50.ga.cel
Reading in : MFP/CCR_T107_MFP_w50.ga.cel
Reading in : MFP/ICRR_T130_MFP_w50.ga.cel
Reading in : MFP/ICRR_T133_MFP_w50.ga.cel
Reading in : MFP/ICRR_T134_MFP_w50.ga.cel
Reading in : MFP/ICRRF_T76_MFP_w50.ga.cel
Reading in : MFP/ICRRF_T77_MFP_w50.ga.cel
Reading in : MFP/ICRRF_T110_MFP_w50.ga.cel
Reading in : MFP/AL_T1_MFP_w80.ga.cel
Reading in : MFP/AL_T3_MFP_w80.ga.cel
Reading in : MFP/AL_T123_MFP_w80.ga.cel
Reading in : MFP/CCR_T33_MFP_w80.ga.cel
Reading in : MFP/CCR_T34_MFP_w80.ga.cel
Reading in : MFP/CCR_T37_MFP_w80.ga.cel
Reading in : MFP/ICRR_T20_MFP_w80.ga.cel
Reading in : MFP/ICRR_T44_MFP_w80.ga.cel
Reading in : MFP/ICRR_T71_MFP_w80.ga.cel
Reading in : MFP/ICRRF_T25_MFP_w80.ga.cel
Reading in : MFP/ICRRF_T116_MFP_w80.ga.cel
Reading in : MFP/ICRRF_T117_MFP_w80.ga.cel
Reading in : Brain/BASELINE_T233_Brain.ga.cel
Reading in : Brain/BASELINE_T234_Brain.ga.cel
Reading in : Brain/BASELINE_T242_Brain.ga.cel
Reading in : Blood/BASELINE_T234_blood.ga.cel
Reading in : Blood/BASELINE_T242_blood.ga.cel
Reading in : Blood/BASELINE_T251_blood.ga.cel
Reading in : MFP/BASELINE_T190_MFP.ga.cel
Reading in : MFP/BASELINE_T193_MFP.ga.cel
Reading in : MFP/BASELINE_T242_MFP.ga.celNormalization
We got a structure with dimensions
Features Samples
292681 69 Now we normalize it using functions from limma
Background correcting
Normalizing
Calculating ExpressionFeatures Samples
36353 69 These are S4 objects, different from typical R objects
We reduced from
Taking just the expression
We only need the expression matrix
[1] 36353 69This is a regular R matrix object
We reduced from 292681 to 36353 rows
Take a look
AL_T3_Brain_w80.ga.cel AL_T67_Brain_w80.ga.cel
14q0_st 0.3875505 0.3659802
14qI-1_st 0.4611885 0.3843568
14qI-1_x_st 0.4611885 0.4297402
14qI-2_st 0.4308127 0.4297402
14qI-3_x_st 0.5527731 0.3664233
14qI-4_st 0.6316946 0.6469497
14qI-4_x_st 0.4580708 0.4804409
14qI-5_st 0.5552395 0.5970109
14qI-6_st -0.0346034 0.2649506
14qI-7_st 0.8116030 0.5584780
14qI-8_st 0.6690536 0.6415562
14qI-8_x_st 0.6538301 0.5694803
14qI-9_x_st 0.2490384 0.5425702
14qII-1_st 0.5560365 0.7858593
14qII-1_x_st 0.5590283 0.6877286Keep only mouse miRNA
We want to use only relevant data
The miRNA description is in another file
annot <- read.csv("miRNA-4_1-st-v1.annotations.20160922.csv", comment = "#")
annot |> subset(Species.Scientific.Name=="Mus musculus") |>
subset(pattern=str_starts("MIMAT")) -> mouse
mouse.data <- norm.Data[mouse$Probe.Set.Name, ]
dim(mouse.data)[1] 3163 69Condition for each sample
In this case each column name includes the condition
[1] AL AL AL CCR CCR CCR ICRR ICRR
[9] ICRR ICRRF ICRRF ICRRF AL AL AL CCR
[17] CCR CCR ICRR ICRR ICRR ICRRF ICRRF ICRRF
[25] AL AL AL CCR CCR CCR ICRR ICRR
[33] ICRR ICRRF ICRRF ICRRF AL AL AL CCR
[41] CCR CCR ICRR ICRR ICRR ICRRF ICRRF ICRRF
[49] AL AL AL CCR CCR CCR ICRR ICRR
[57] ICRR ICRRF ICRRF ICRRF BASELINE BASELINE BASELINE BASELINE
[65] BASELINE BASELINE BASELINE BASELINE BASELINE
Levels: AL BASELINE CCR ICRR ICRRFFitting the linear model
With the description of condition, we build a design matrix
AL BASELINE CCR ICRR ICRRF
1 1 0 0 0 0
2 1 0 0 0 0
3 1 0 0 0 0
4 0 0 1 0 0
5 0 0 1 0 0
6 0 0 1 0 0
7 0 0 0 1 0
8 0 0 0 1 0
9 0 0 0 1 0
10 0 0 0 0 1
11 0 0 0 0 1
12 0 0 0 0 1
13 1 0 0 0 0
14 1 0 0 0 0
15 1 0 0 0 0
16 0 0 1 0 0
17 0 0 1 0 0
18 0 0 1 0 0
19 0 0 0 1 0
20 0 0 0 1 0
21 0 0 0 1 0
22 0 0 0 0 1
23 0 0 0 0 1
24 0 0 0 0 1
25 1 0 0 0 0
26 1 0 0 0 0
27 1 0 0 0 0
28 0 0 1 0 0
29 0 0 1 0 0
30 0 0 1 0 0
31 0 0 0 1 0
32 0 0 0 1 0
33 0 0 0 1 0
34 0 0 0 0 1
35 0 0 0 0 1
36 0 0 0 0 1
37 1 0 0 0 0
38 1 0 0 0 0
39 1 0 0 0 0
40 0 0 1 0 0
41 0 0 1 0 0
42 0 0 1 0 0
43 0 0 0 1 0
44 0 0 0 1 0
45 0 0 0 1 0
46 0 0 0 0 1
47 0 0 0 0 1
48 0 0 0 0 1
49 1 0 0 0 0
50 1 0 0 0 0
51 1 0 0 0 0
52 0 0 1 0 0
53 0 0 1 0 0
54 0 0 1 0 0
55 0 0 0 1 0
56 0 0 0 1 0
57 0 0 0 1 0
58 0 0 0 0 1
59 0 0 0 0 1
60 0 0 0 0 1
61 0 1 0 0 0
62 0 1 0 0 0
63 0 1 0 0 0
64 0 1 0 0 0
65 0 1 0 0 0
66 0 1 0 0 0
67 0 1 0 0 0
68 0 1 0 0 0
69 0 1 0 0 0
attr(,"assign")
[1] 1 1 1 1 1
attr(,"contrasts")
attr(,"contrasts")$condition
[1] "contr.treatment"Making contrasts
Fitting the model
Evaluating p-values
CvsA RvsA RFvsA RvsC RFvsC RvsRF AveExpr F P.Value
MIMAT0014941_st -0.22 -0.27 0.16 -0.05 0.38 -0.43 0.66 8.04 0.00
MI0009961_st 0.19 0.14 0.39 -0.06 0.20 -0.25 1.50 4.89 0.00
MI0022894_st 0.00 0.09 0.28 0.09 0.28 -0.19 0.87 4.87 0.00
MI0004300_st -0.09 0.20 -0.09 0.29 0.00 0.29 0.82 4.30 0.01
MI0000560_st 0.01 0.11 0.34 0.09 0.33 -0.23 0.99 4.24 0.01
MIMAT0014950_st -0.04 0.10 0.22 0.14 0.25 -0.12 0.83 4.00 0.01
MI0022778_st -0.04 -0.05 0.11 -0.02 0.15 -0.17 0.55 3.99 0.01
MIMAT0014822_st -0.14 0.23 0.01 0.37 0.14 0.22 0.76 3.88 0.01
MI0022921_st 0.01 -0.05 0.21 -0.06 0.20 -0.26 0.74 3.83 0.01
MIMAT0014838_st -0.12 -0.07 0.12 0.05 0.25 -0.20 0.43 3.82 0.01
adj.P.Val
MIMAT0014941_st 0.39
MI0009961_st 1.00
MI0022894_st 1.00
MI0004300_st 1.00
MI0000560_st 1.00
MIMAT0014950_st 1.00
MI0022778_st 1.00
MIMAT0014822_st 1.00
MI0022921_st 1.00
MIMAT0014838_st 1.00(in this example the p-values are bad, but anyway)
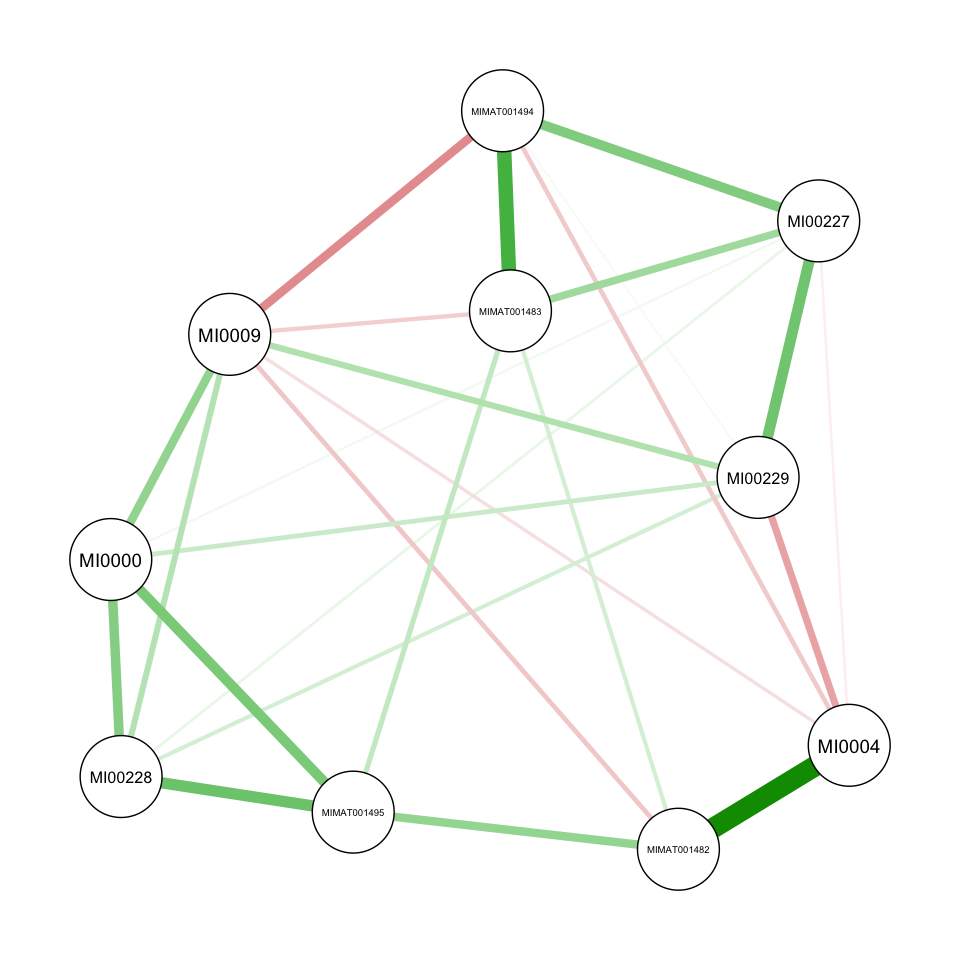
Experimental covariance
The last 4 columns are not differential expression values, so we drop them
Now we calculate the covariance between the rows, so we transpose top
Using glasso
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 16.00 -0.77 -4.55 0.67 -4.58 -4.14 -4.03 0.00 -4.38 -6.30
[2,] -0.77 17.29 -1.87 3.84 -3.31 0.00 0.00 0.82 -3.23 0.00
[3,] -4.55 -1.87 23.50 0.00 -3.09 0.00 0.00 0.00 -0.56 0.00
[4,] 0.67 3.84 0.00 20.81 0.00 0.00 0.00 -3.18 0.58 0.00
[5,] -4.58 -3.31 -3.09 0.00 20.76 -1.44 0.00 0.00 -2.10 -1.03
[6,] -4.14 0.00 0.00 0.00 -1.44 26.80 0.00 0.00 0.00 0.00
[7,] -4.03 0.00 0.00 0.00 0.00 0.00 31.51 0.00 0.00 0.00
[8,] 0.00 0.82 0.00 -3.18 0.00 0.00 0.00 19.69 0.00 0.00
[9,] -4.38 -3.23 -0.56 0.58 -2.10 0.00 0.00 0.00 23.59 0.00
[10,] -6.30 0.00 0.00 0.00 -1.03 0.00 0.00 0.00 0.00 25.06We observe that the pseudoinverse has many zeros, as expected
Graphical view
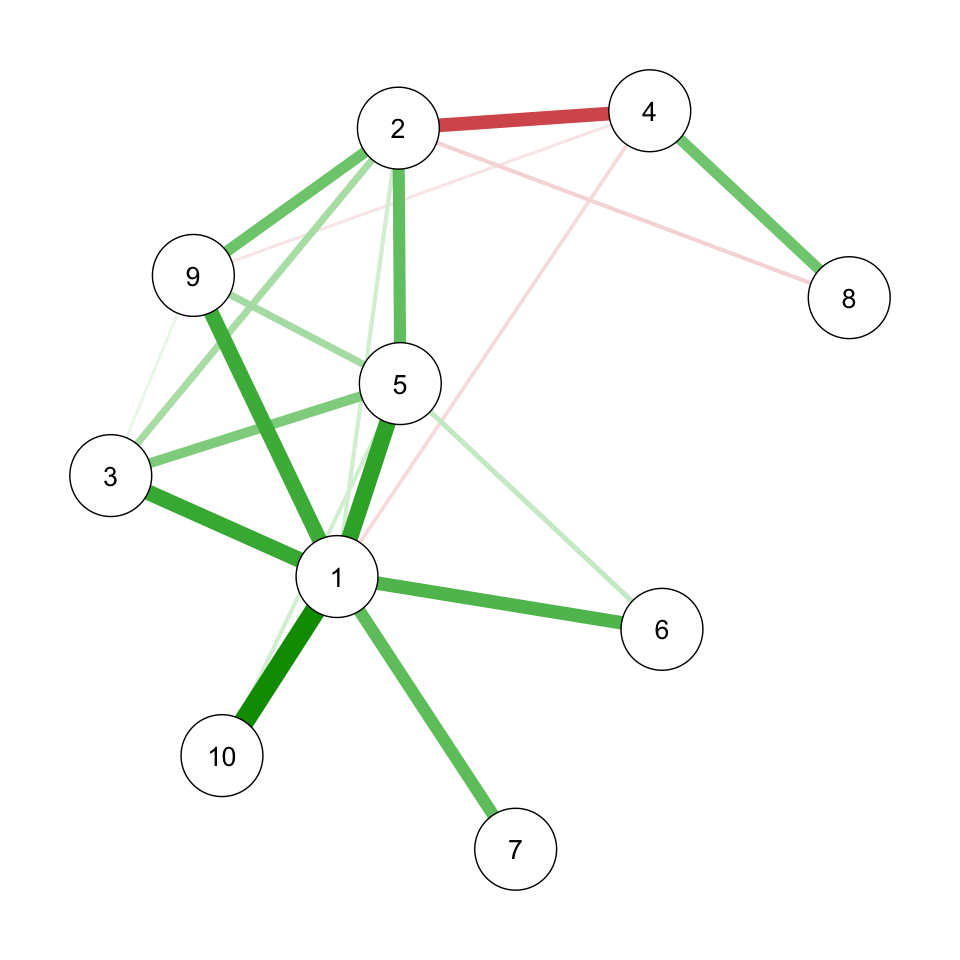
Optimal lambda/rho