Class 9: Finding the replication origin
Computing for Molecular Biology 2
Andrés Aravena, PhD
2 April 2021
Finding the
Replication Origin
in Bacteria
Finding the replication origin
In Bacteria there is only one replication origin
- The GC skew is positive in one part of chromosome
- And negative on the other, on average
- It changes sign in two places
The reason for this asymmetry is the DNA replication process
The origin of replication is in one of the sign changing places
How can we find the change of sign
We want to write a program so we can find the replication origin on many different genomes
We can answer questions like:
- Are both replichores of the same size?
- If they are different, what is the biological reason?
- Is there some insertion?
- What are the extra genes? What do they do?
Statistics in parts of the genome
Local statistics look only through a small window
A Window is a part of the genome, from an initial position, and with a fixed size
For example, we can analyze a region of 500 bp starting at position 3000 of the genome
Evaluate gc_skew() window by window
We traverse the genome with several windows of fixed size
Each window starts after the end of the previous one
In other words, the windows do not overlap
Calculate GC skew in a window
We created this function in Class 6. This is a new version
window_GC_skew <- function(position, size, dna) {
V <- toupper(dna[seq(from=position, length.out=size)])
count_C <- sum(V=="C")
count_G <- sum(V=="G")
if( (count_C+count_G) == 0) {
return(0)
} else {
return( (count_G-count_C)/(count_C+count_G) )
}
}This new code does the same as the old one.
Can you see how does it work?
Create a vector with all window’s positions
We start at the first DNA position, until the last.
Be careful to not step out of DNA
Now we apply window_gc_skew() to each element of win_pos
The result depends on window’s size
Result for size=1E5
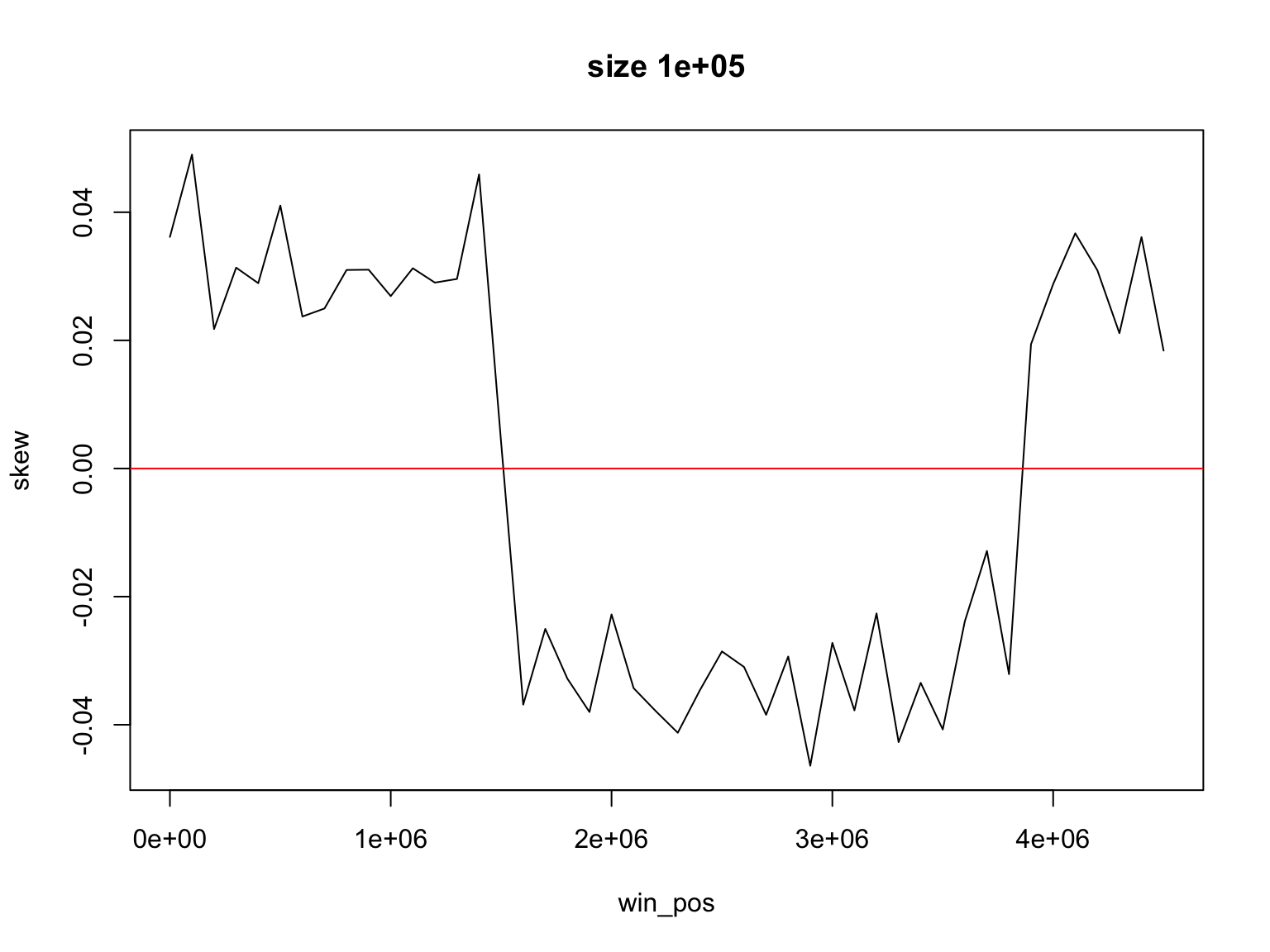
We look for the change of sign
We find two places where GC-skew changes sign
[1] 1500000 3800000but the window size is 1E5, so the origin is somewhere between
[1] 1450000 1550000or
[1] 3750000 3850000(how would you find the place where GC skew changes its sign?)
Let’s use smaller size
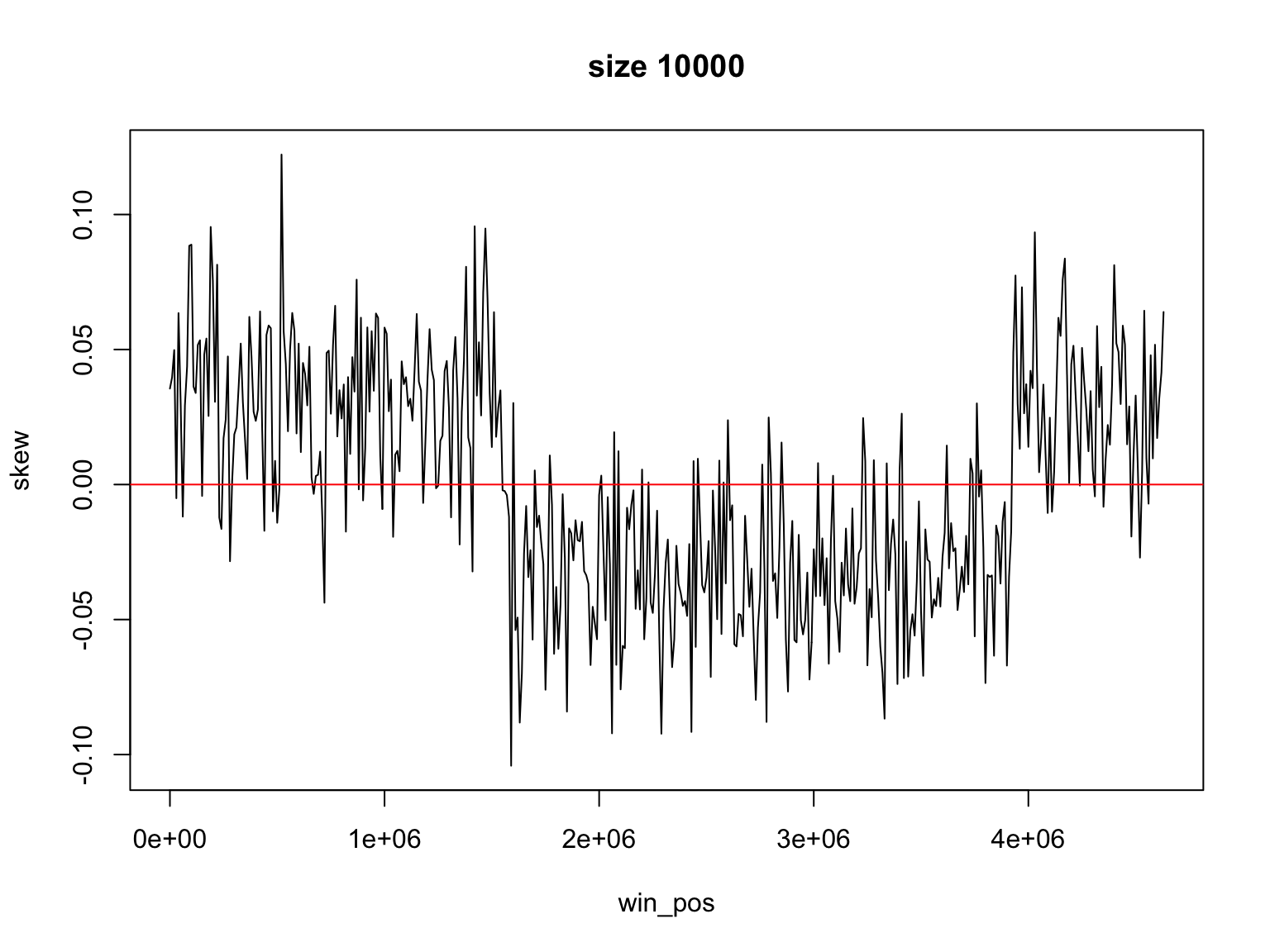
Result for size=1E3
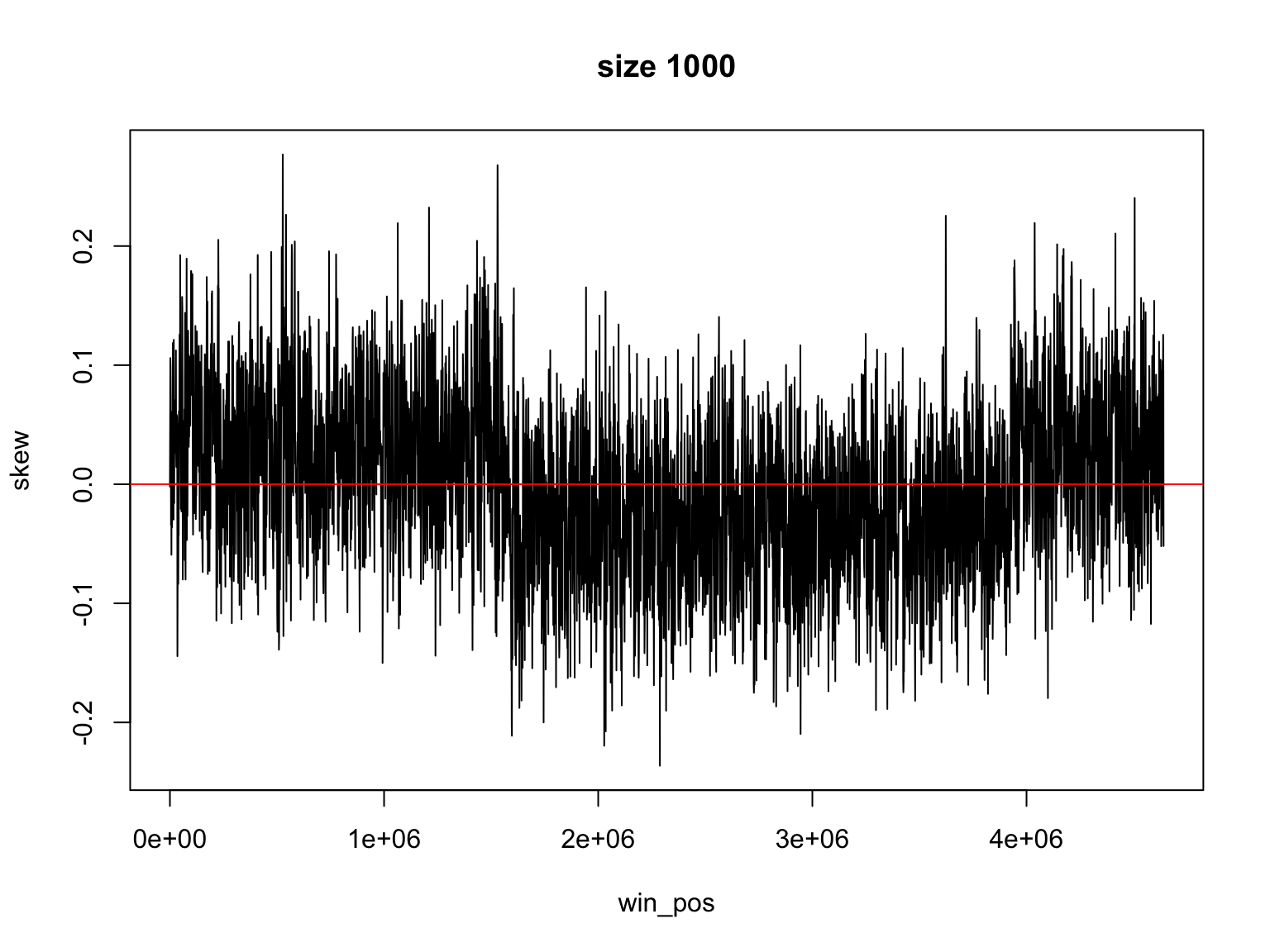
Result for size=1E2

It is not so easy
Finding the change of sign is not so easy
There is a trade off
Large windows have large margin of error
Smaller windows give values that change too much
skew data is noisy
But most of times it has the same sign
What happens if we sum the skew values?
Positives will be more positive,
negatives will be more negative
Finding where
the sign changes
There is an easier way
Instead of looking directly at skew, we can look at accumulative skew
which can also be written as
Calculating Accumulative Sums
We have skew, we want acc_skew
Now, instead of the change of sign, we look for the minimum and maximum
(Why?)
The dictionary says
- accumulative
- § (adjective) increasing in quantity, degree, or force by successive additions.
- § increasing, accumulative, growing, mounting; collective, aggregate, amassed.
- § the accumulative effect of two years of drought
- § the effects of pollution are accumulative
Now max & min show the change of sign

Finding where is
the largest value
Position of the maximum
The maximum value of acc_skew is
[1] 4.9877It is not really important
We are looking for position, not value of maximum
Position of the maximum
This is the index of the maximum in the acc_skew vector
[1] 155This is always an integer number, between 1 and length(acc_skew)
It is not the position in the genome
Position of the maximum
Since acc_skew vector has the same length as win_pos, we can find the genomic position of the maximum with this code
[1] 1540001win_pos contains the genome positions of each window
This code gives us the position of the window with maximum GC skew
Locating origin depends on window size
The largest GC skew is at
The replication origin is located in the interval between
place_of_max - size/2place_of_max + size/2
[1] 1535000 1545000Skew and Accumulative skew
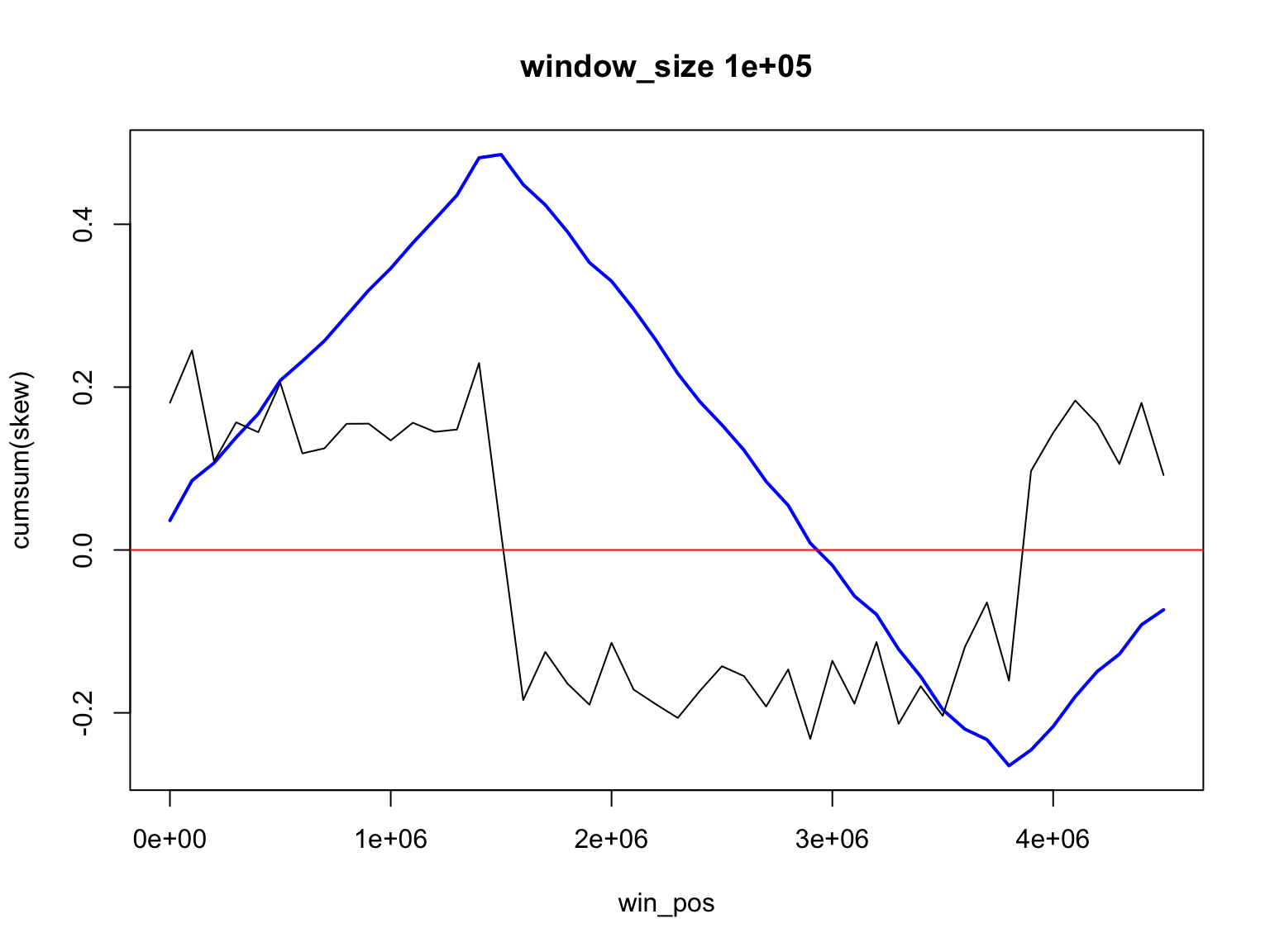
Skew and Accumulative skew
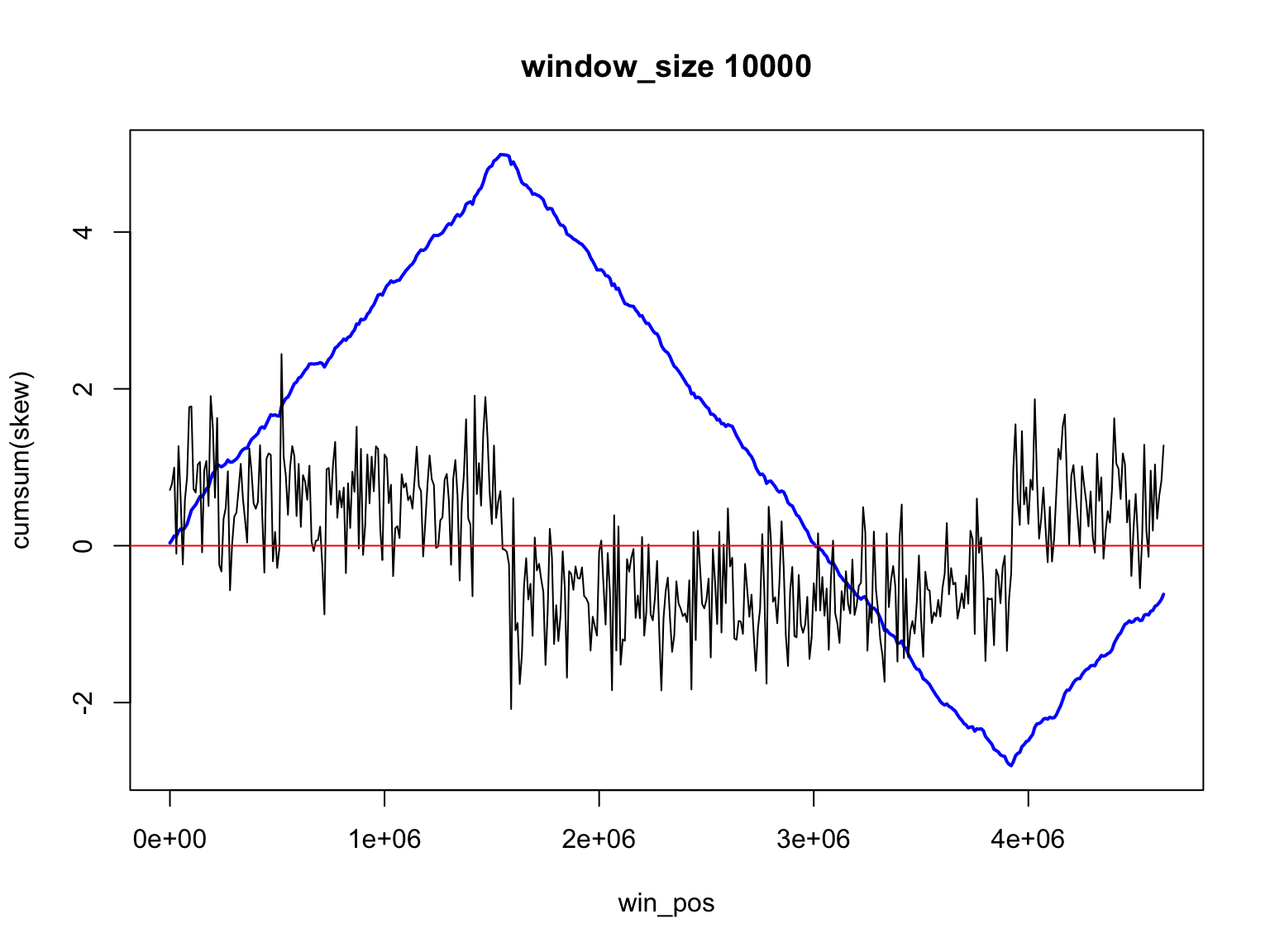
Skew and Accumulative skew
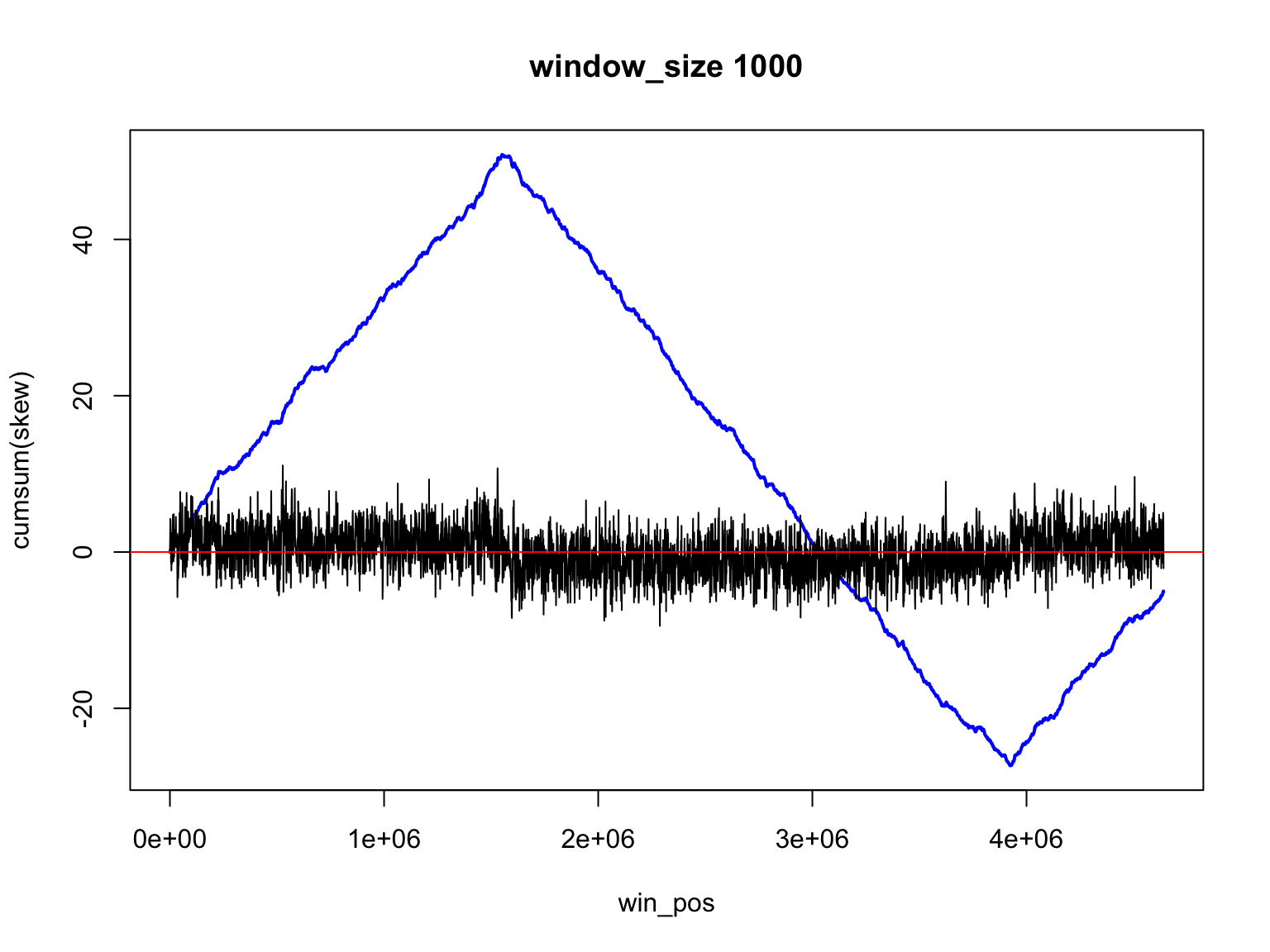
skew changes fast, acc_skew changes slow
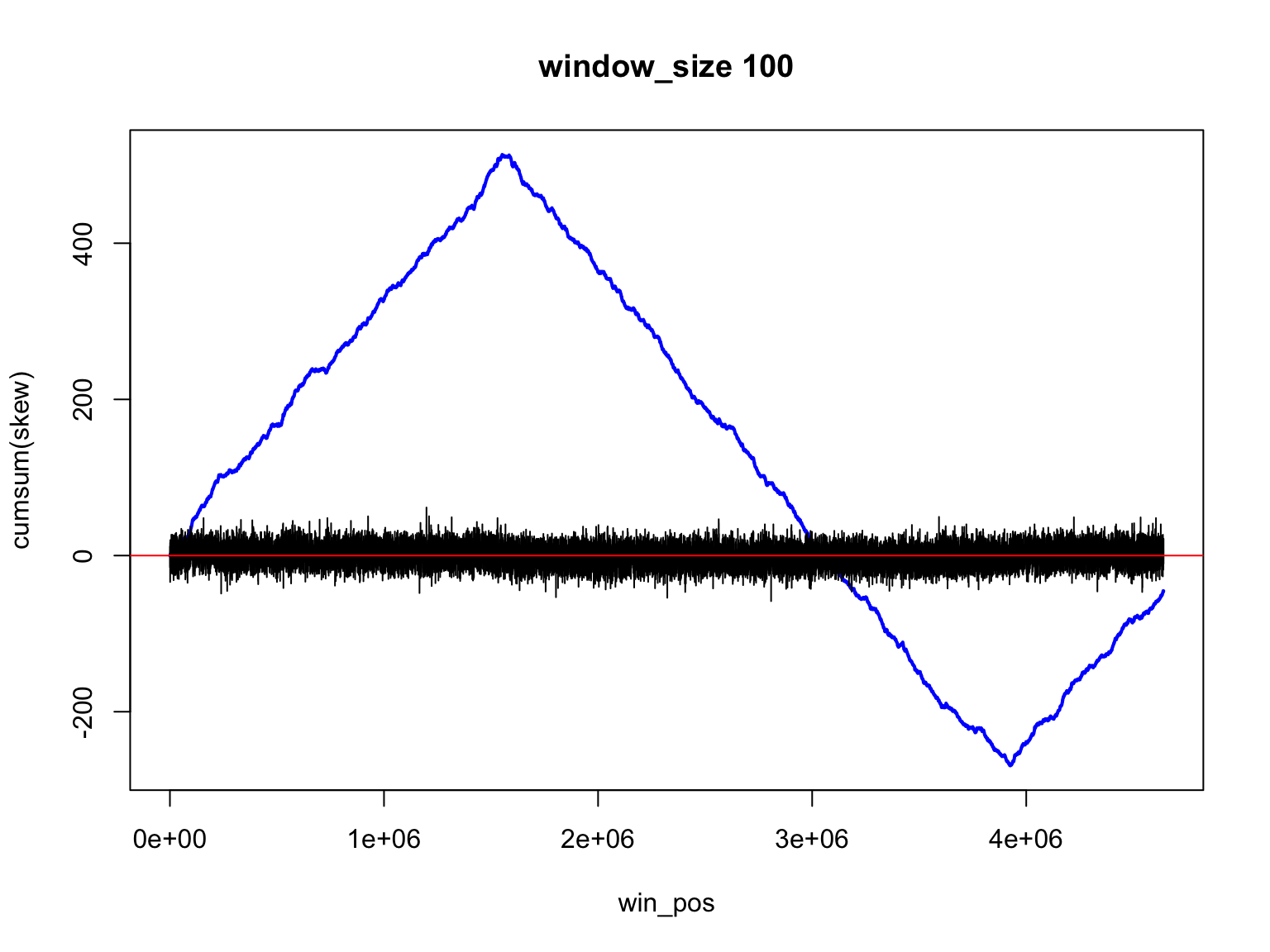
Accumulative sums
Accumulative sum
Everyday you spend some money
Some days you receive money
How much money you have each day?
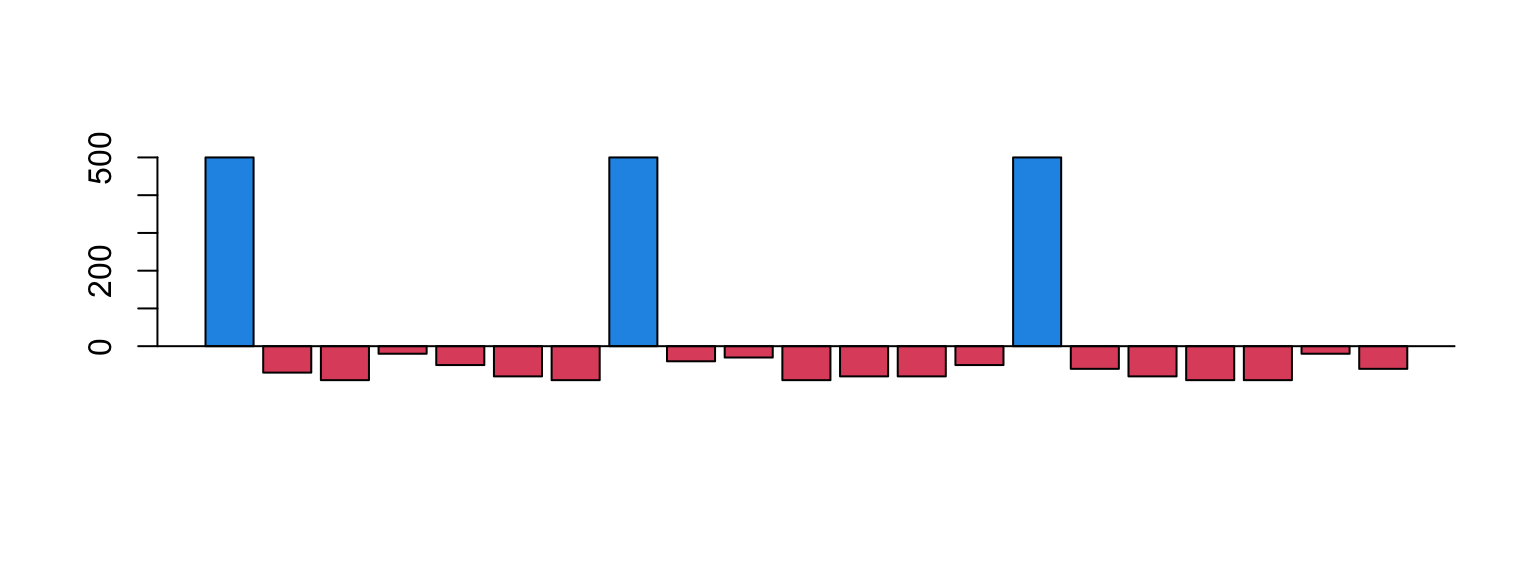
Accumulative Sum: daily balance
The income can be positive (salary) or negative (expenses)
The balance on first day is your initial money
The balance of today is the balance of yesterday plus the income of today
balance and income are vectors
Since they have different values every day, we represent the income on day i by income[i]
We also represent the balance on day i by balance[i]
The vector income has the values for all days
The element income[i] has the value for a single day
Balance looks like this
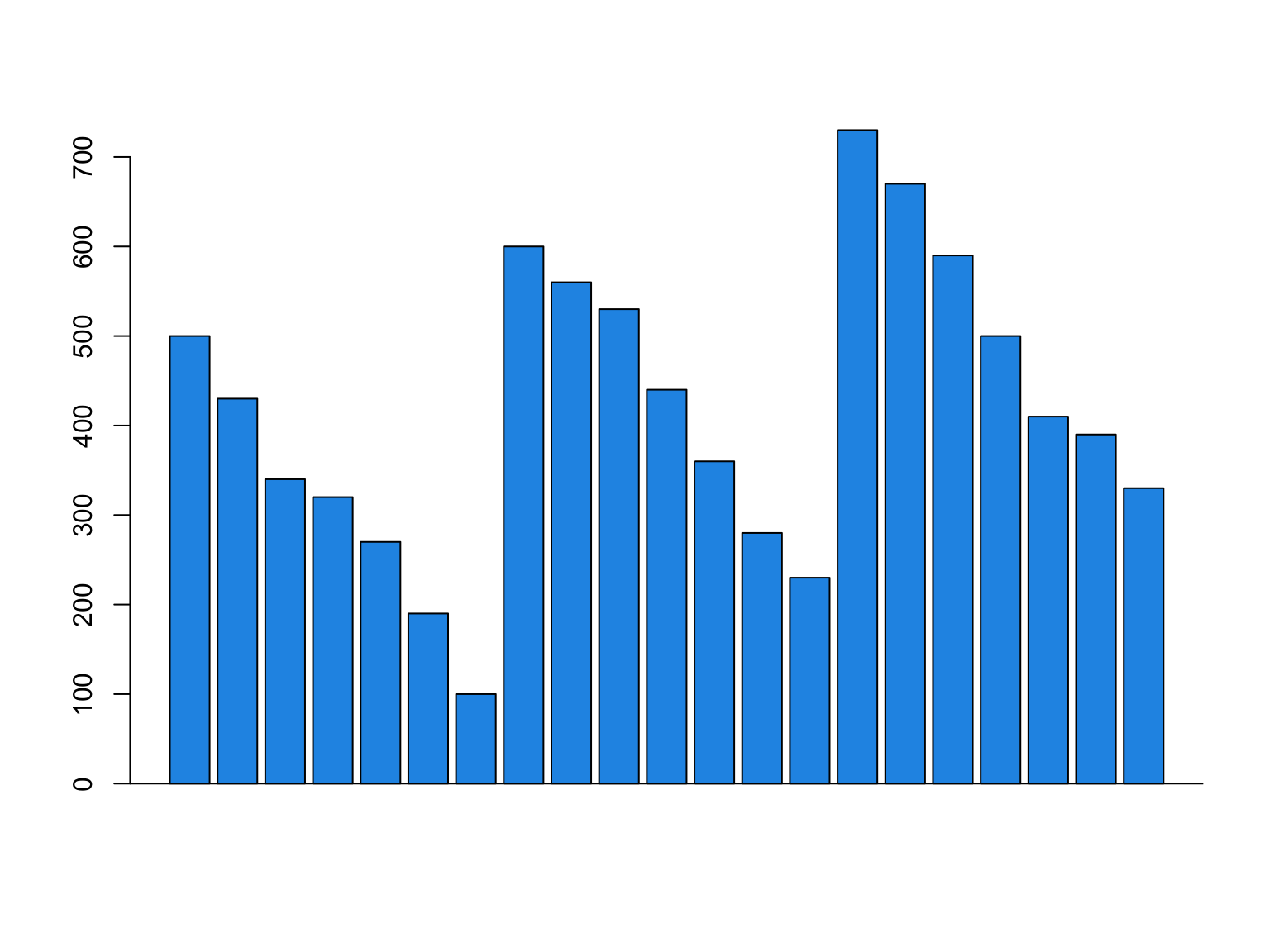
Accumulative sums are useful
If we know the income, we can get balance
If we know the skew, we can get acc_skew
If we know the change, we can know the state
Exercise
Write your own version of cumsum