Class 5: Practice with sapply. The FOR loop
Computing for Molecular Biology 2
Andrés Aravena, PhD
19 March 2021
Finding patterns and using them
In the previous class we learned about patterns
When we see that several parts are very similar, we can forget about details and write a generic part
We write the generic part inside a function, that we can reuse several times
Functions: code in a box
Function are like black boxes
To use a function we give the input and we receive the output

Functions are contracts between the person who uses and the person who writes the function
Appply the function to many things
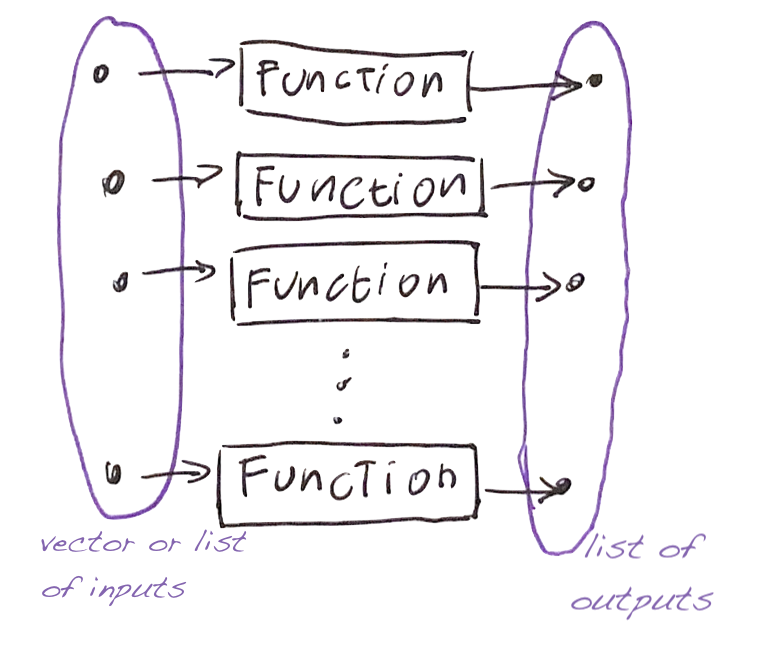
The command
- takes a vector (or list)
v, - applies the function
funto each element, - and returns a list.
Mapping a vector into another
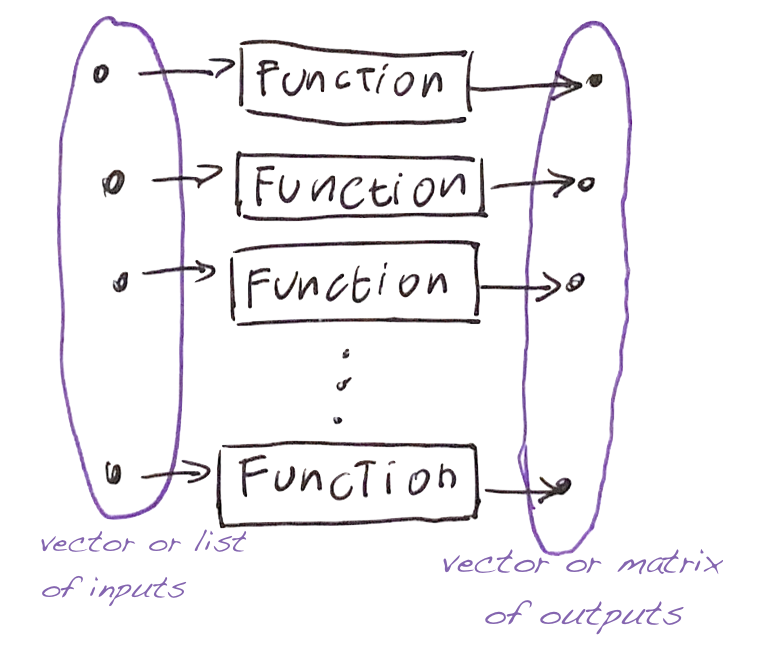
The command
- takes a vector (or list)
v, - applies the function
funto each element, - and tries to return a vector (or a matrix).
Common case: sequence to vector
The most common case is when the vector of inputs is a numeric sequence, like
There is another way
In previous version of this course we used for to repeat the same commands several times
The parts are
- an index
- a vector (or list) of values for the index
- commands that depend on the index, wraped in
{}
Most common usage of for(){}
The parts are
- an index
- start value for the index
- end value for the index
- commands that depend on the index, wraped in
{}
The world for is special, like function or sequence
Application:
Do all genes have the same GC content?
Exercise
Is the GC content uniform through all genome?
Do all genes have the same GC content?
What function do we need to answer this question?
What is the name, the inputs, and the output?
From previous class
We need to read E.coli genes into a list
library(seqinr)
genes <- read.fasta("NC_000913.ffn", seqtype = "DNA", set.attributes = FALSE)
length(genes)[1] 4140If all is right, we get 4140 genes.
The GC content function
We will use the same function we created in the previous class
GC for all genes
Let’s calculate GC content of all genes without using sapply()
This works, but we need to store the results somewhere
Notice that a for loop does not give any value.
In contrast, lapply and sapply always give a result.
Storing the results
This code does not work
Error in eval(expr, envir, enclos): object 'answer' not foundWe cannot assign to answer[i]
unless the vector answer exists before
We must create the vector answer before using it
The complete code is this
Notice that length(genes) is used twice
- in
rep(NA, length(genes))to create the vector - in the range of the
for(){}loop
sapply() is much easier
Instead of worrying about creating a vector and assign each value to it, we can use sapply()
This is faster to write and faster to execute. It can do several steps at the same time
It gives the same result
[1] TRUEWhich one shall we use?
Having several options gives you great power
And with great power comes great responsibility
Use sapply in most cases, if the order of steps is not important
Use for loops when
- the new result depends on the previous results
- you want to test an idea step-by-step
- (sometimes) they are easier to understand
Application: GC skew
GC Skew
What is the size of \(G-C\) respect to the total \(G+C\)?
Calculate the ratio \[\frac{G-C}{G+C}\] for each gene and plot it
What do you see?
GC skew
There are more G than C in the leading strand
There are more C than G in the lagging strand
GC skew changes sign at the boundaries of the two replichores
This corresponds to DNA replication origin or terminus
First: Write it in English
(or in any human language)
- download genes of E.coli
- For each gene
count C and G
calculate
(G-C) / (G+C)return the calculated value
Second: write a function
We can adapt a similar function, like gene_GC_content_v3
calculate_GC_skew <- function(dna) {
V <- toupper(dna)
count_C <- sum(V=="C")
count_G <- sum(V=="G")
GC_skew <- (count_G-count_C)/(count_C+count_G)
return(GC_skew)
}We can test it some basic cases
[1] -0.2941176Third: sapply the function
This is interesting
In bacteria, we expect to see that GC skew changes sign between leading and lagging strand
We did not saw that change of sign in the plot
- Is it true that the GC skew changes sign?
- If so, how can we explain this result?
We answer these questions on the next class
