Class 7: Affine Gaps are more realistic
Bioinformatics
Andrés Aravena
October 18, 2021
Global, Semi-Global and Local alignment
What is the difference?
When do we use each one?
Substitution Scoring Matrix
Example for DNA
A C G T
A 1 -2 -2 -2
C -2 1 -2 -2
G -2 -2 1 -2
T -2 -2 -2 1Score of changing “A” for “C” is -2
Changing “A” for “A” (i.e. preserving) has score +1
PAM30 matrix
A R N D C Q E G H I L K M F P S T W
A 6 -7 -4 -3 -6 -4 -2 -2 -7 -5 -6 -7 -5 -8 -2 0 -1 -13
R -7 8 -6 -10 -8 -2 -9 -9 -2 -5 -8 0 -4 -9 -4 -3 -6 -2
N -4 -6 8 2 -11 -3 -2 -3 0 -5 -7 -1 -9 -9 -6 0 -2 -8
D -3 -10 2 8 -14 -2 2 -3 -4 -7 -12 -4 -11 -15 -8 -4 -5 -15
C -6 -8 -11 -14 10 -14 -14 -9 -7 -6 -15 -14 -13 -13 -8 -3 -8 -15
Q -4 -2 -3 -2 -14 8 1 -7 1 -8 -5 -3 -4 -13 -3 -5 -5 -13
E -2 -9 -2 2 -14 1 8 -4 -5 -5 -9 -4 -7 -14 -5 -4 -6 -17
G -2 -9 -3 -3 -9 -7 -4 6 -9 -11 -10 -7 -8 -9 -6 -2 -6 -15
H -7 -2 0 -4 -7 1 -5 -9 9 -9 -6 -6 -10 -6 -4 -6 -7 -7
I -5 -5 -5 -7 -6 -8 -5 -11 -9 8 -1 -6 -1 -2 -8 -7 -2 -14
L -6 -8 -7 -12 -15 -5 -9 -10 -6 -1 7 -8 1 -3 -7 -8 -7 -6
K -7 0 -1 -4 -14 -3 -4 -7 -6 -6 -8 7 -2 -14 -6 -4 -3 -12
M -5 -4 -9 -11 -13 -4 -7 -8 -10 -1 1 -2 11 -4 -8 -5 -4 -13
F -8 -9 -9 -15 -13 -13 -14 -9 -6 -2 -3 -14 -4 9 -10 -6 -9 -4
P -2 -4 -6 -8 -8 -3 -5 -6 -4 -8 -7 -6 -8 -10 8 -2 -4 -14
S 0 -3 0 -4 -3 -5 -4 -2 -6 -7 -8 -4 -5 -6 -2 6 0 -5
T -1 -6 -2 -5 -8 -5 -6 -6 -7 -2 -7 -3 -4 -9 -4 0 7 -13
W -13 -2 -8 -15 -15 -13 -17 -15 -7 -14 -6 -12 -13 -4 -14 -5 -13 13
Y -8 -10 -4 -11 -4 -12 -8 -14 -3 -6 -7 -9 -11 2 -13 -7 -6 -5
V -2 -8 -8 -8 -6 -7 -6 -5 -6 2 -2 -9 -1 -8 -6 -6 -3 -15
B -3 -7 6 6 -12 -3 1 -3 -1 -6 -9 -2 -10 -10 -7 -1 -3 -10
J -6 -7 -6 -10 -9 -5 -7 -10 -7 5 6 -7 0 -2 -7 -8 -5 -7
Z -3 -4 -3 1 -14 6 6 -5 -1 -6 -7 -4 -5 -13 -4 -5 -6 -14
X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
* -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17 -17
Y V B J Z X *
A -8 -2 -3 -6 -3 -1 -17
R -10 -8 -7 -7 -4 -1 -17
N -4 -8 6 -6 -3 -1 -17
D -11 -8 6 -10 1 -1 -17
C -4 -6 -12 -9 -14 -1 -17
Q -12 -7 -3 -5 6 -1 -17
E -8 -6 1 -7 6 -1 -17
G -14 -5 -3 -10 -5 -1 -17
H -3 -6 -1 -7 -1 -1 -17
I -6 2 -6 5 -6 -1 -17
L -7 -2 -9 6 -7 -1 -17
K -9 -9 -2 -7 -4 -1 -17
M -11 -1 -10 0 -5 -1 -17
F 2 -8 -10 -2 -13 -1 -17
P -13 -6 -7 -7 -4 -1 -17
S -7 -6 -1 -8 -5 -1 -17
T -6 -3 -3 -5 -6 -1 -17
W -5 -15 -10 -7 -14 -1 -17
Y 10 -7 -6 -7 -9 -1 -17
V -7 7 -8 0 -6 -1 -17
B -6 -8 6 -8 0 -1 -17
J -7 0 -8 6 -6 -1 -17
Z -9 -6 0 -6 6 -1 -17
X -1 -1 -1 -1 -1 -1 -17
* -17 -17 -17 -17 -17 -17 1BLOSUM matrices
Using local alignment we can identify conserved regions
In 1992 Steven Henikoff and Jorja Henikoff created new substitution matrices based on local alignment of blocks
BLOcks SUbstitution Matrix
Idea: each protein domain can evolve at different speeds
BLOSUM62
A R N D C Q E G H I L K M F P S T W Y V B J Z X
A 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -1 -1 -2 -1 1 0 -3 -2 0 -2 -1 -1 -1
R -1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 -1 -2 0 -1
N -2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 4 -3 0 -1
D -2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 4 -3 1 -1
C 0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 -3 -1 -3 -1
Q -1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 -1 -2 0 -2 4 -1
E -1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 1 -3 4 -1
G 0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 -1 -4 -2 -1
H -2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -1 -2 -1 -2 -2 2 -3 0 -3 0 -1
I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 -3 3 -3 -1
L -1 -2 -3 -4 -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 -2 -1 1 -4 3 -3 -1
K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2 0 -3 1 -1
M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 -1 -1 1 -3 2 -1 -1
F -2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1 -3 0 -3 -1
P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2 -2 -3 -1 -1
S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2 0 -2 0 -1
T 0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5 -2 -2 0 -1 -1 -1 -1
W -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11 2 -3 -4 -2 -2 -1
Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1 -3 -1 -2 -1
V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 -3 2 -2 -1
B -2 -1 4 4 -3 0 1 -1 0 -3 -4 0 -3 -3 -2 0 -1 -4 -3 -3 4 -3 0 -1
J -1 -2 -3 -3 -1 -2 -3 -4 -3 3 3 -3 2 0 -3 -2 -1 -2 -1 2 -3 3 -3 -1
Z -1 0 0 1 -3 4 4 -2 0 -3 -3 1 -1 -3 -1 0 -1 -2 -2 -2 0 -3 4 -1
X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
* -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4
*
A -4
R -4
N -4
D -4
C -4
Q -4
E -4
G -4
H -4
I -4
L -4
K -4
M -4
F -4
P -4
S -4
T -4
W -4
Y -4
V -4
B -4
J -4
Z -4
X -4
* 1Homework
Read th paper by M. O. Dayhoff and R. M. Schwartz, Chapter 22: A model of evolutionary change in proteins, in Atlas of Protein Sequence and Structure, 1978.
Copy the matrix in Figure 80, and from that create the matrices on Figures 82-84
Show me the formulas you used
Deadline: next Monday
Looking the diagonals
For local alignment we have two cases
- Ungapped, where we can only move in diagonal
- Gapped, where we can move diagonal, horizontal, and vertical
- Horizontal and vertical movement gives gaps
Global and semi-global alignments always have gaps
Gaps
Gap penalty
We prefer this alignment
GGGTAACCTACCTC
||| ||||| ||||
GGGCAACCTGCCTCinstead of this other alignment
GGGT-AACCTA-CCTC
||| ||||| ||||
GGG-CAACCT-GCCTCThus, gap penalty must be greater than mismatch penalty
Long gaps instead of short ones
We prefer this alignment
TCAAAGAG---GATA
||| ||| ||||
TCA--GAGGGGGATAinstead of this other alignment
TCAAAGA-G-G-ATA
|| | || | | |||
TC-A-GAGGGGGATAWe want few long gaps instead of many short gaps
Affine gaps
Gap values must reflect how real insertions and deletions occur in nature
We observe that, once an indel event starts, it can easily grow
If the polymerase jumps, then it can jump a long distance
To represent this, we use affine gaps
Linear v/s affine
So far we considered only linear gaps
The penalty of \(n\) consecutive gaps is \(n\cdot G\)
(\(G\) is the gap penalty)
Now we consider affine gaps, where the first gap is expensive, but the consecutive are cheap
The penalty of \(n\) consecutive gaps is \(I + n\cdot E\)
\(I\) is the initial gap penalty, \(E\) is the gap extension penalty
Traceback
Traceback
After we built the matrix, we must go back from the “optimal score” finding which was the path
There may be more than one solution
Some programs build the alignment at the same time they build the matrix, but that requires more memory
Example
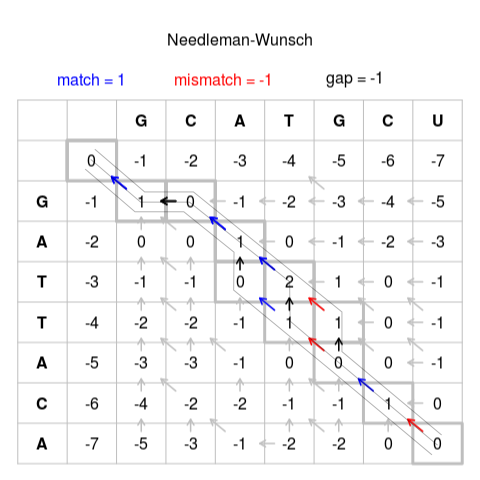
Solution 1
GCAT-GCU
G-ATTACASolution 2
GCA-TGCU
G-ATTACASolution 3
GCATG-CU
G-ATTACA