- Solve simple problems
- Decomposition
- Pattern matching
- Abstraction
- Systems Analysis
- Identify the system parts and processes
- Use R to simulate a system
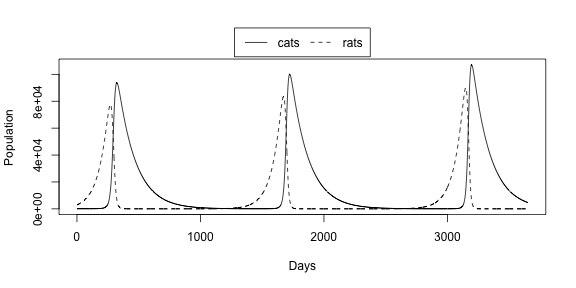
- See what happens in the long term
for(i in 2:N) {
r1 <- r1_rate*H[i-1]*H[i-1]*O[i-1]
r2 <- r2_rate*W[i-1]
d_W[i] <- r1 - r2
d_O[i] <- -r1 + r2
d_H[i] <- -2*r1 + 2*r2
W[i] <- W[i-1] + d_W[i]
O[i] <- O[i-1] + d_O[i]
H[i] <- H[i-1] + d_H[i]
}