Class 26: Drawing beautiful plots
Computing for Molecular Biology 1
Andrés Aravena, PhD
11 January 2021
Grammar of Graphics
Grammar
Grammar is:
How to speak and make sense
For example, if we say
make how speak and to sense
we have the same words, but they do not make sense
English grammar
In English we say
John eats fruit
- There is a subject: “John”
- There is a verb: “eats”
- There may be an adverb: “slowly”
- There is a predicate: “fruit”
- There may be an adjective: “fresh”
John slowly eats fresh fruit
Grammar of graphics
- what is the data
- which columns map to each graphic attributes
- what statistical transformation are needed
- which geometric object will be draw
- what last-minute position adjustment is needed
There are many explanations on the web. Google them
We use ggplot2
There was an old “ggplot” package. We use the second version.
This package gives us many commands. The basic ones are
ggplot()geom_point(),geom_line(), and othergeom_…commands- Several
scale_x_…,scale_y_…, and otherscale_…commands theme
First plot
Warning: Removed 18 rows containing missing values (geom_point).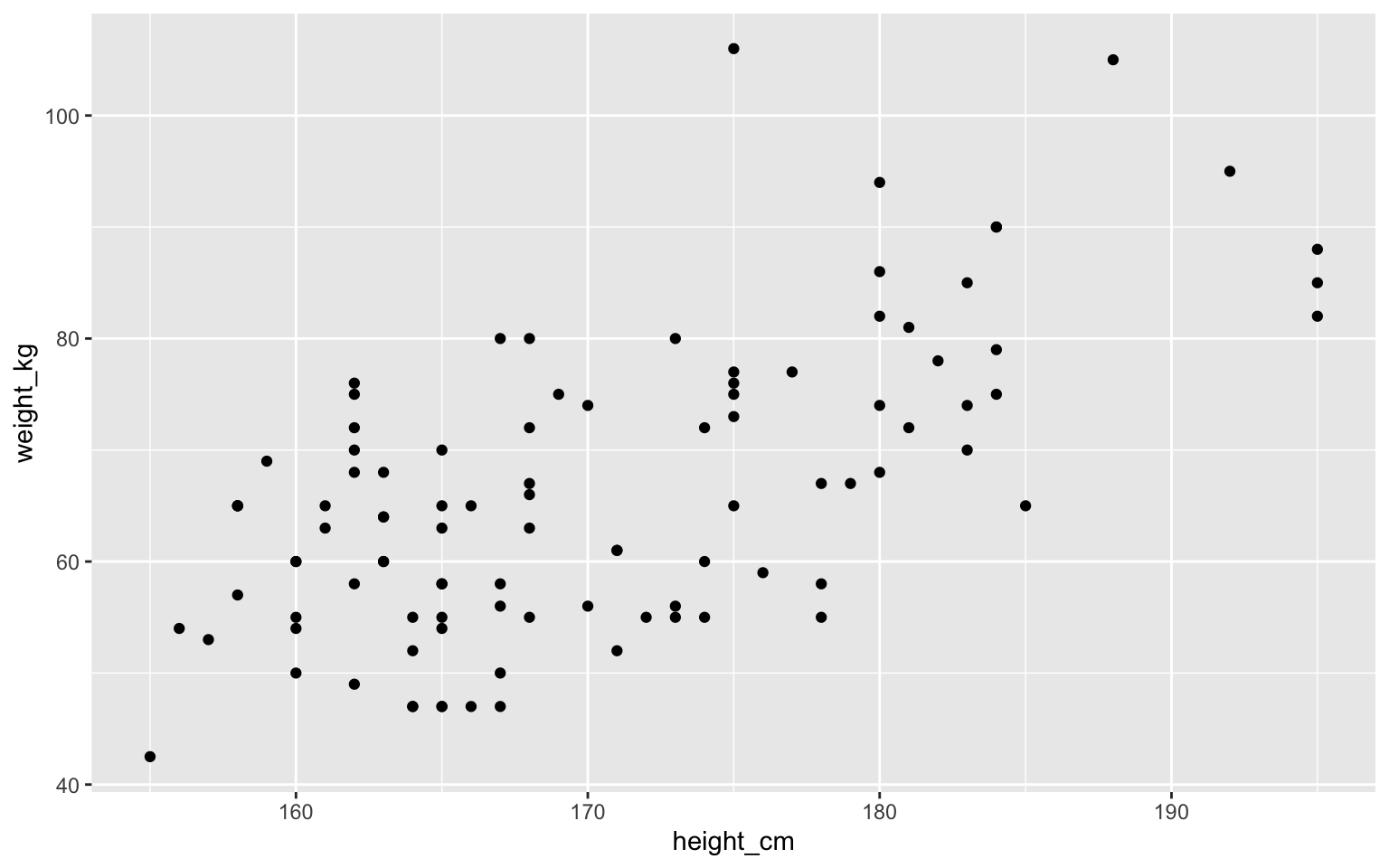
Parts of this command
- We start with
ggplot(). It takes two inputs- a data frame, in this case
students - an
aes()object, called aesthetic map
- a data frame, in this case
aes()describe how data frame columns are mapped to visual properties (aesthetics)- in this case we chose the
x=andy=parts
- in this case we chose the
- Finally, we add the geometry to draw the points
- In this case we use
geom_point()
- In this case we use
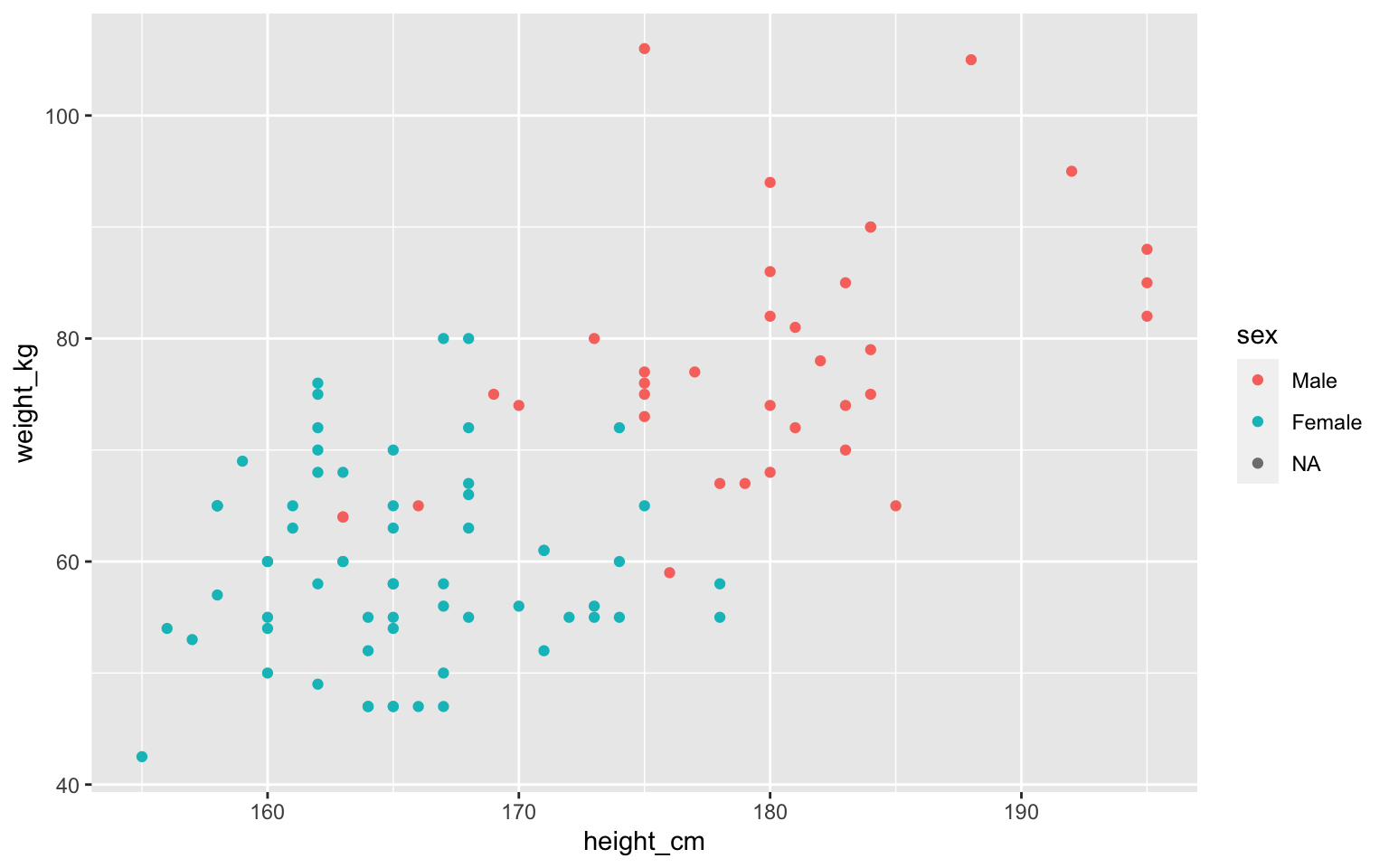
Aesthetics: how to show each column
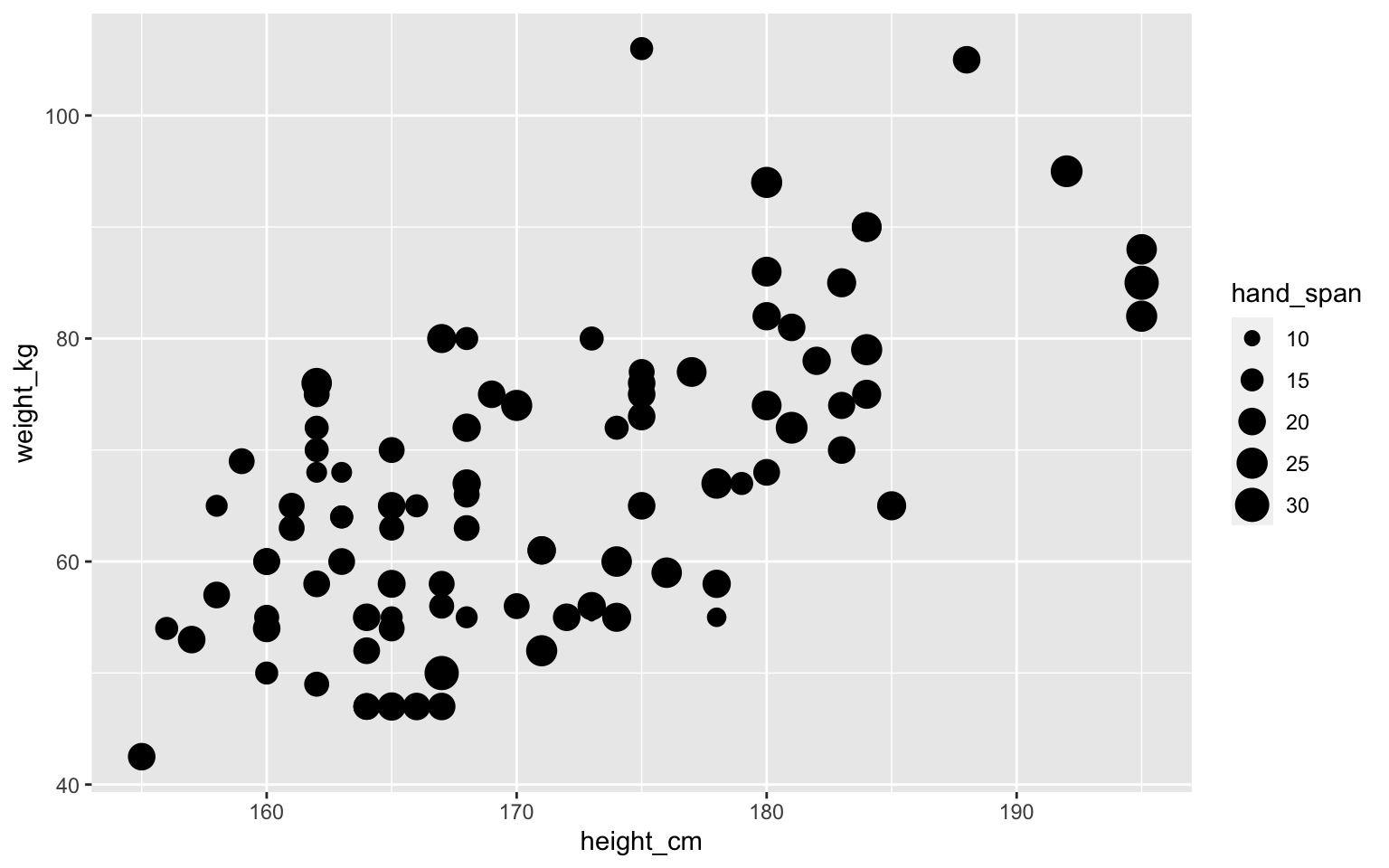
Point size can show some data
Transparent points using alpha
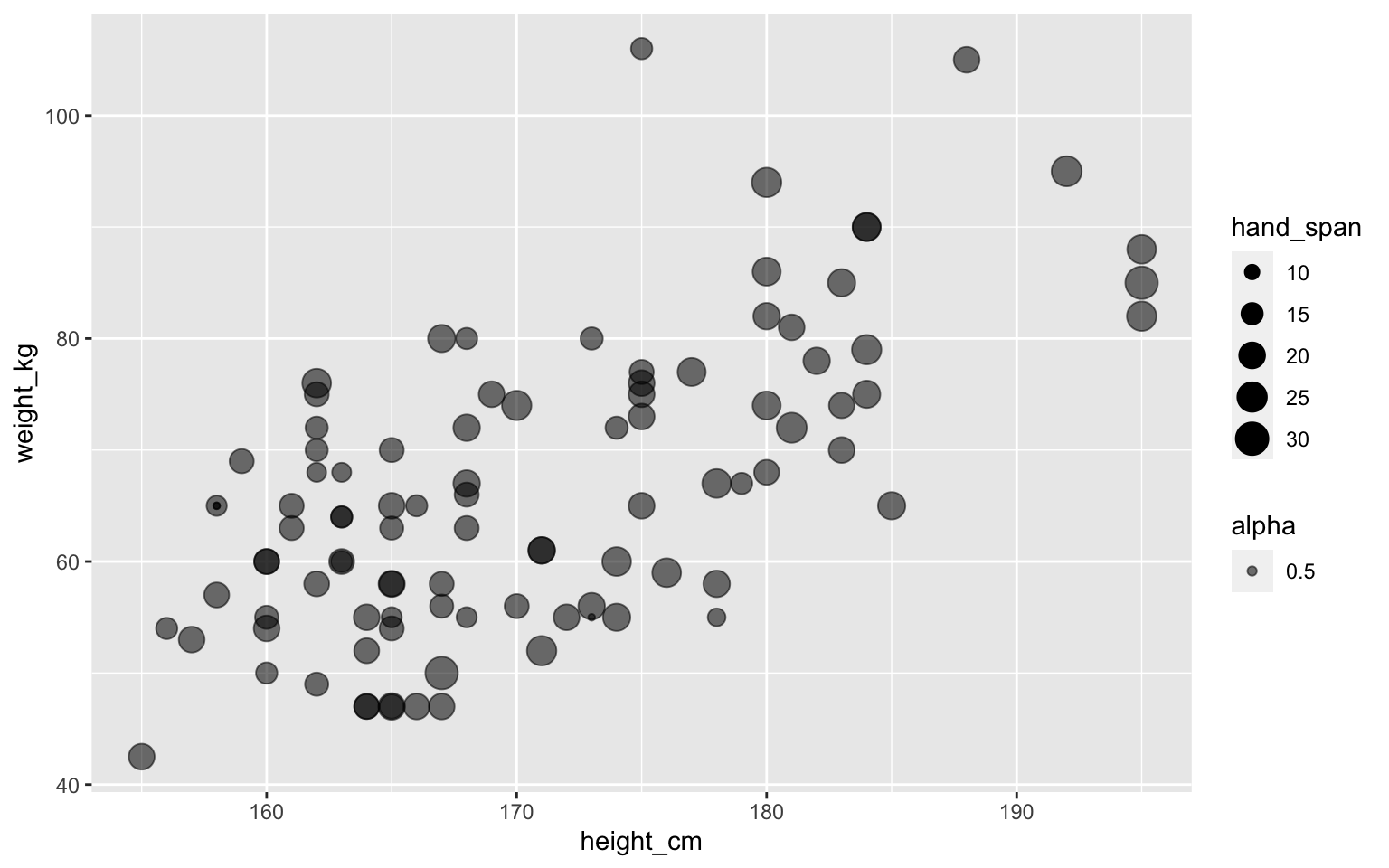
We can put aesthetics in geom_point()
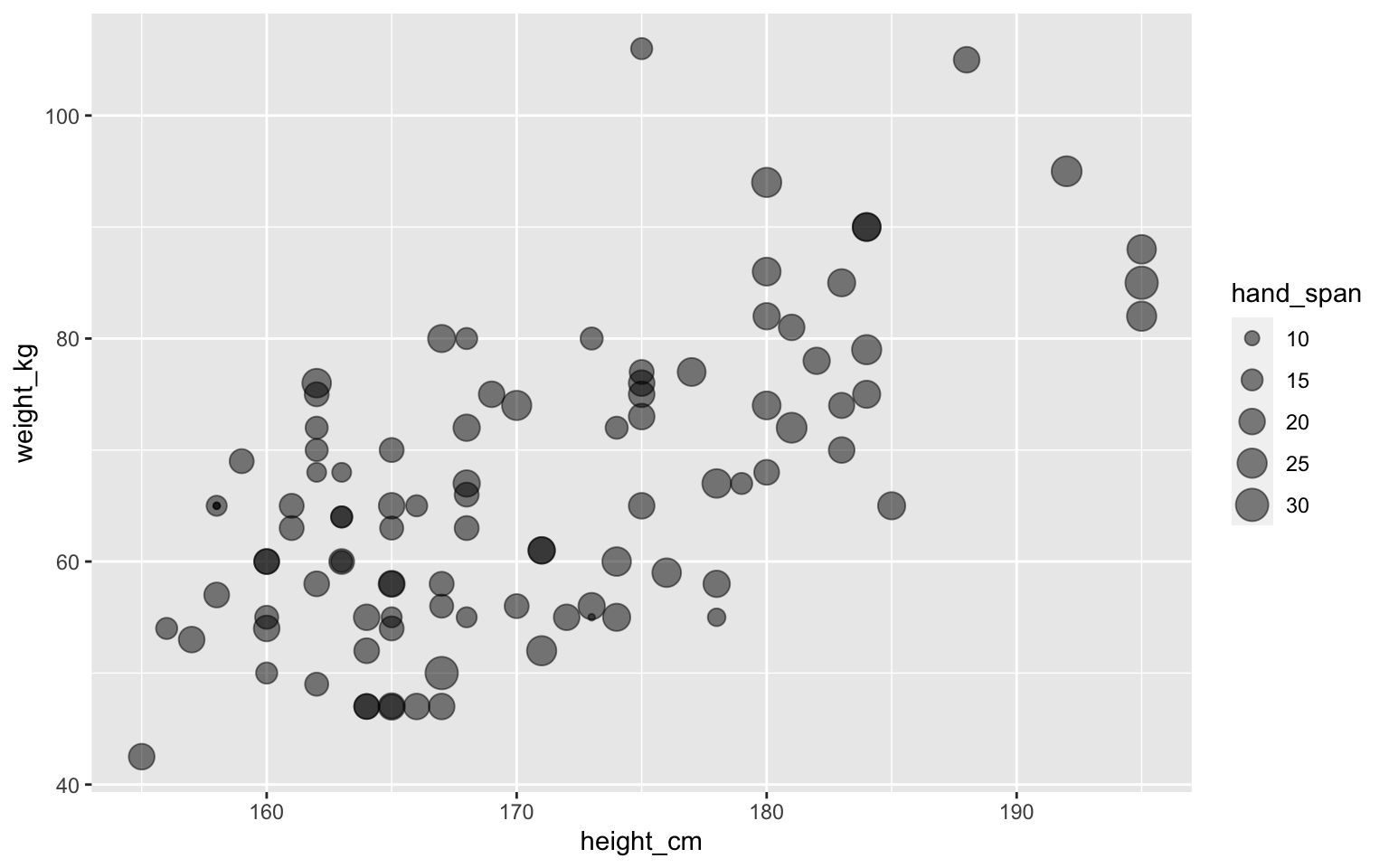
Assigning multiple aesthetics
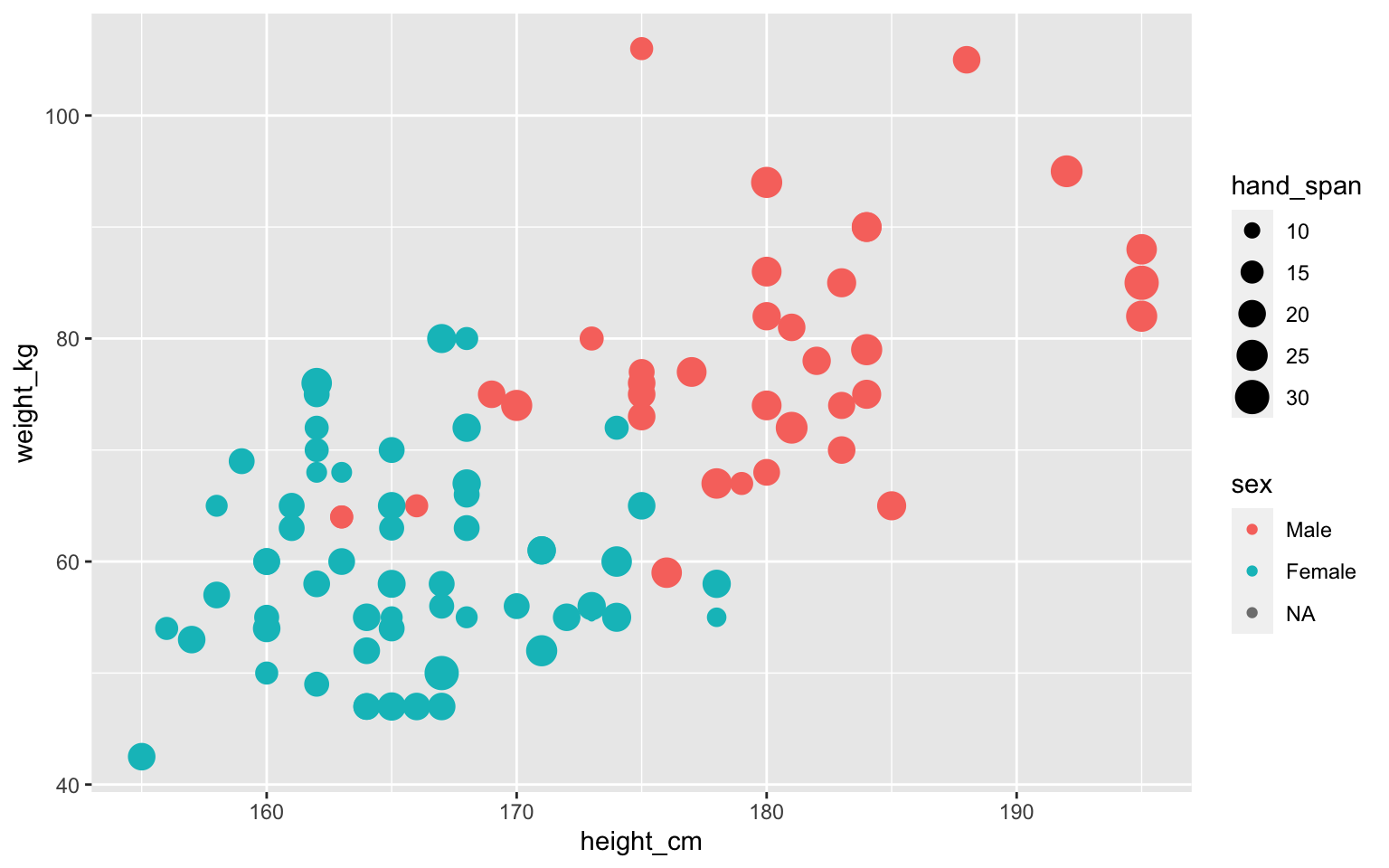
Some shapes have color and fill
ggplot(students, aes(x=height_cm, y=weight_kg,
size=hand_span, fill=sex)) +
geom_point(alpha=0.5, shape="circle filled", color="black")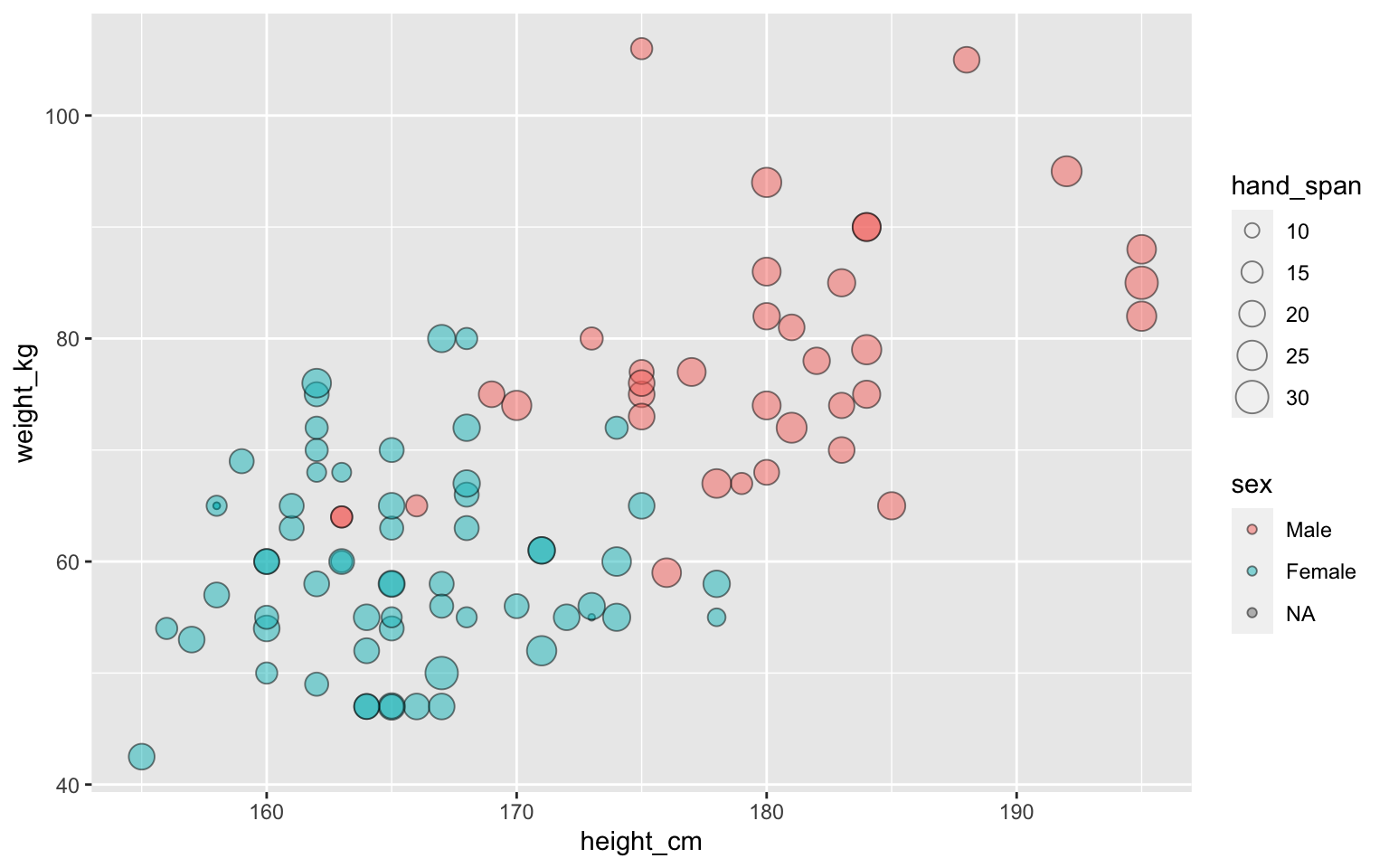
Boxplot
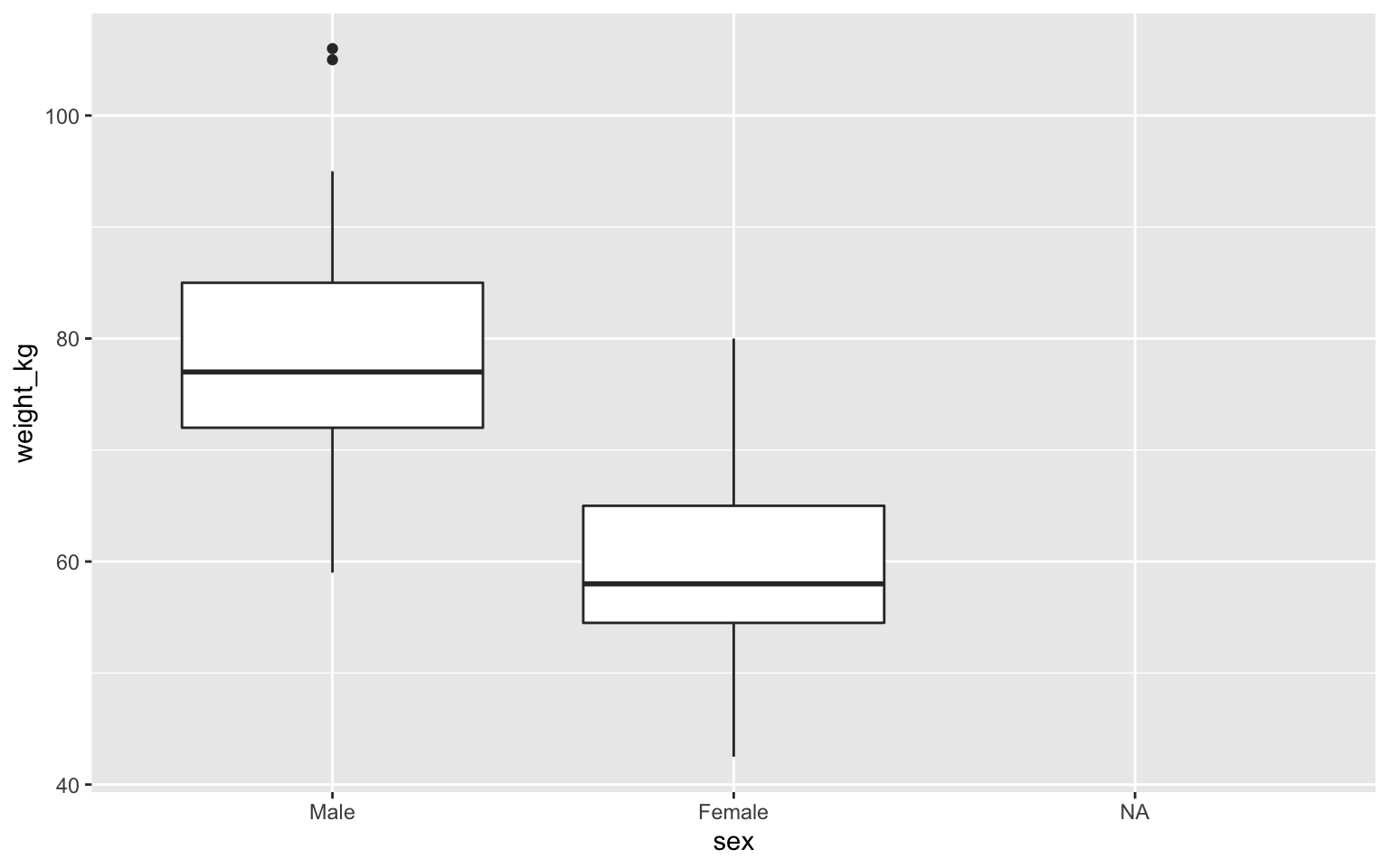
Boxplot with color
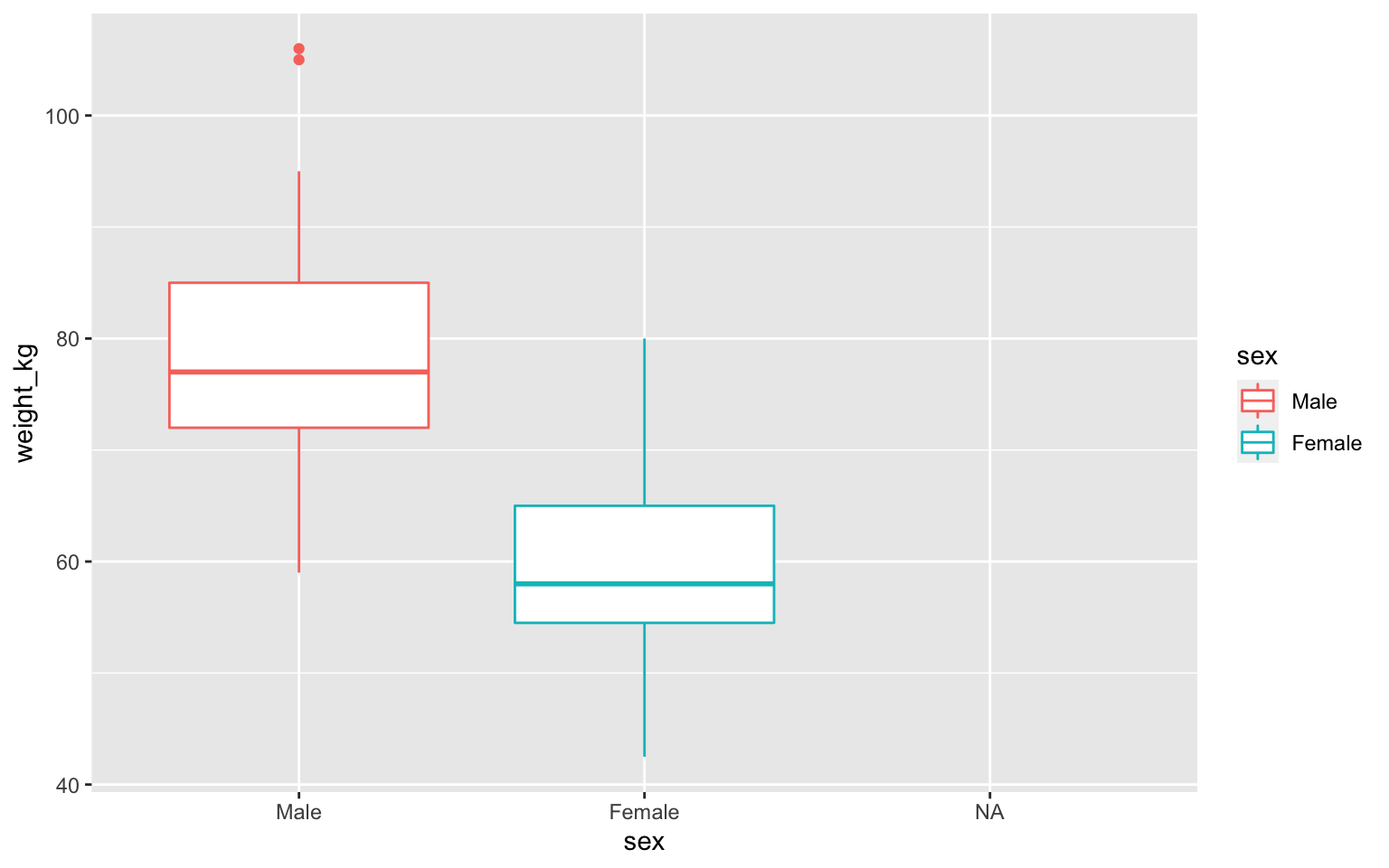
Boxplot with fill
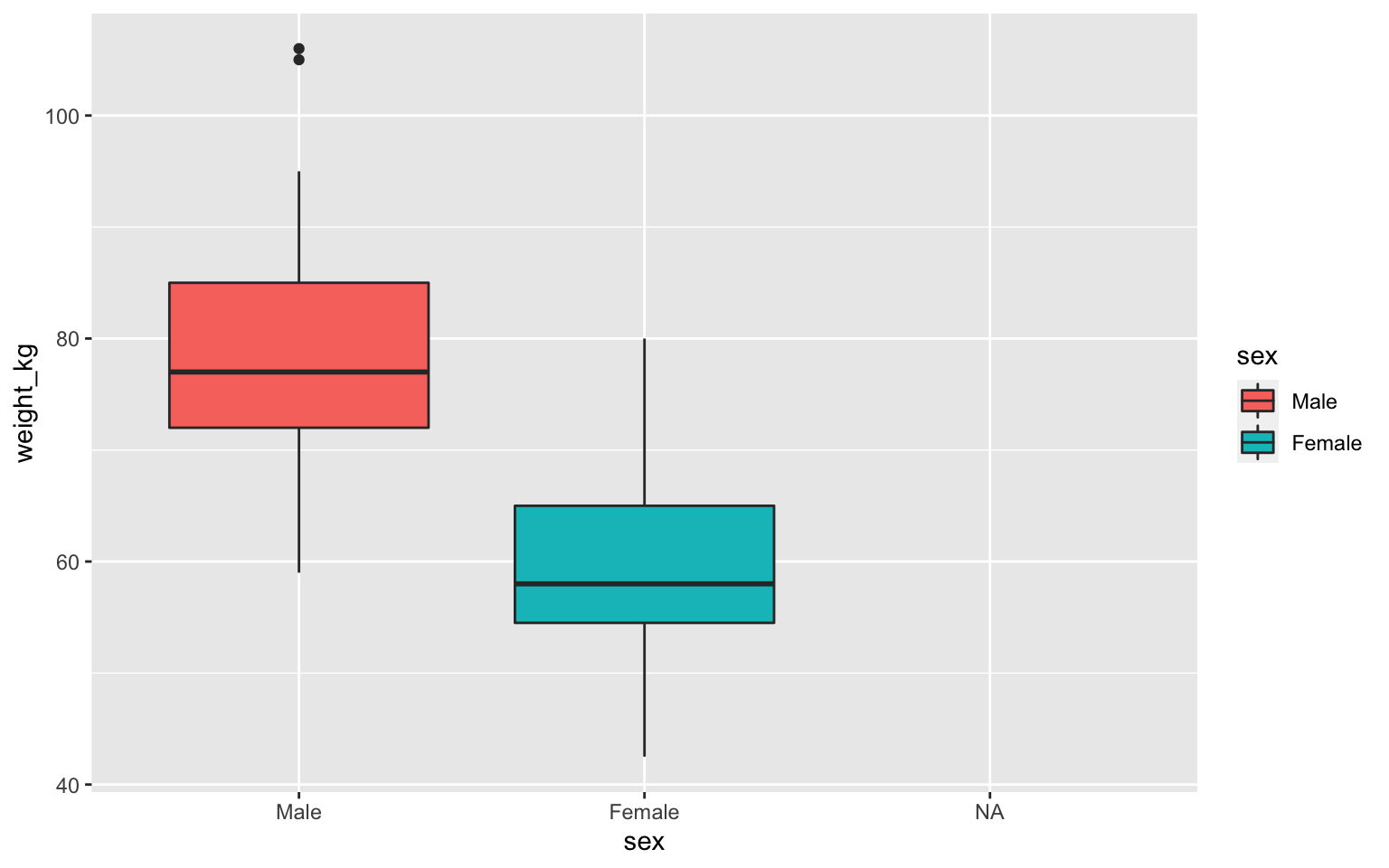
Plotting a single column
Barplot counts values in one column
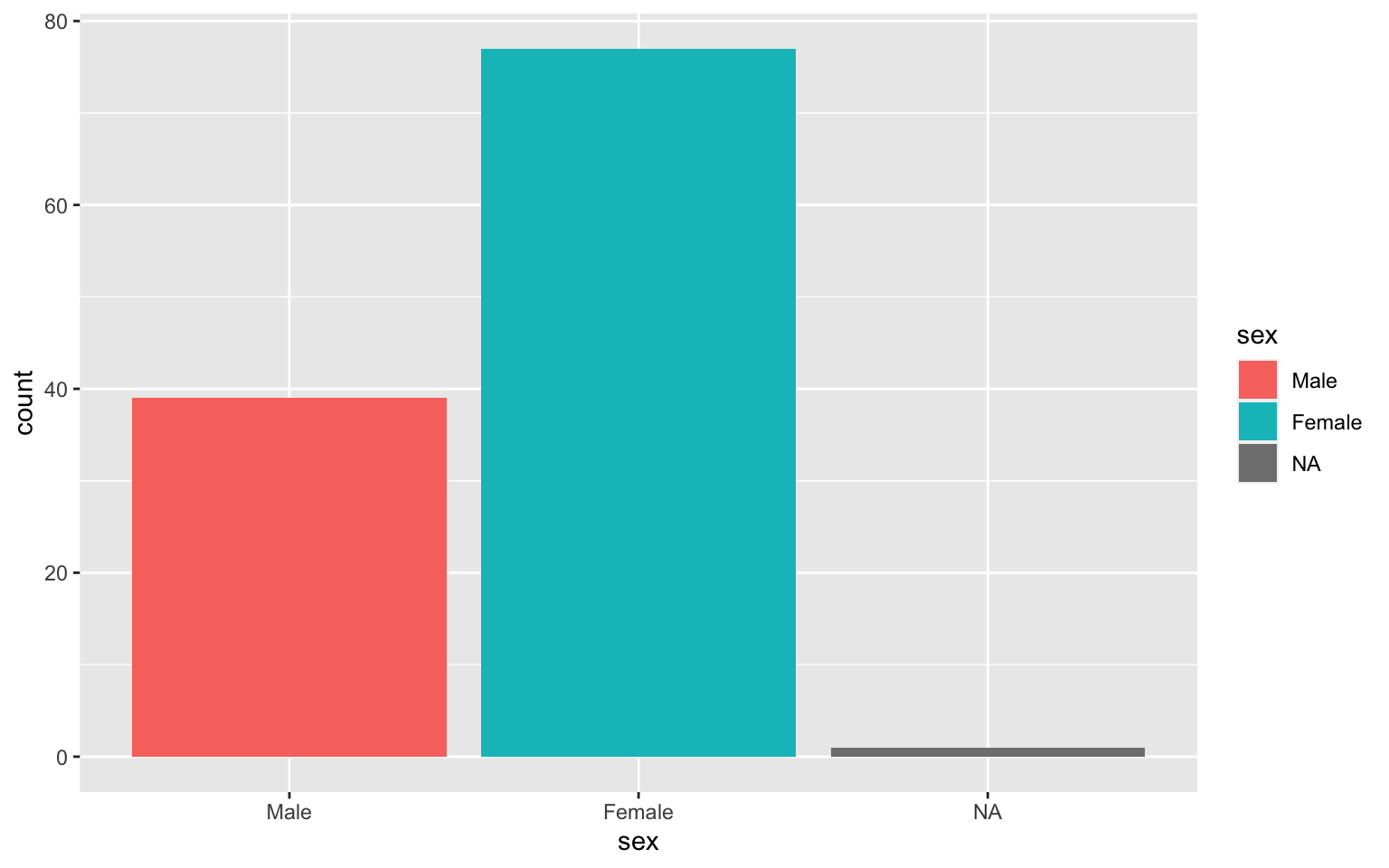
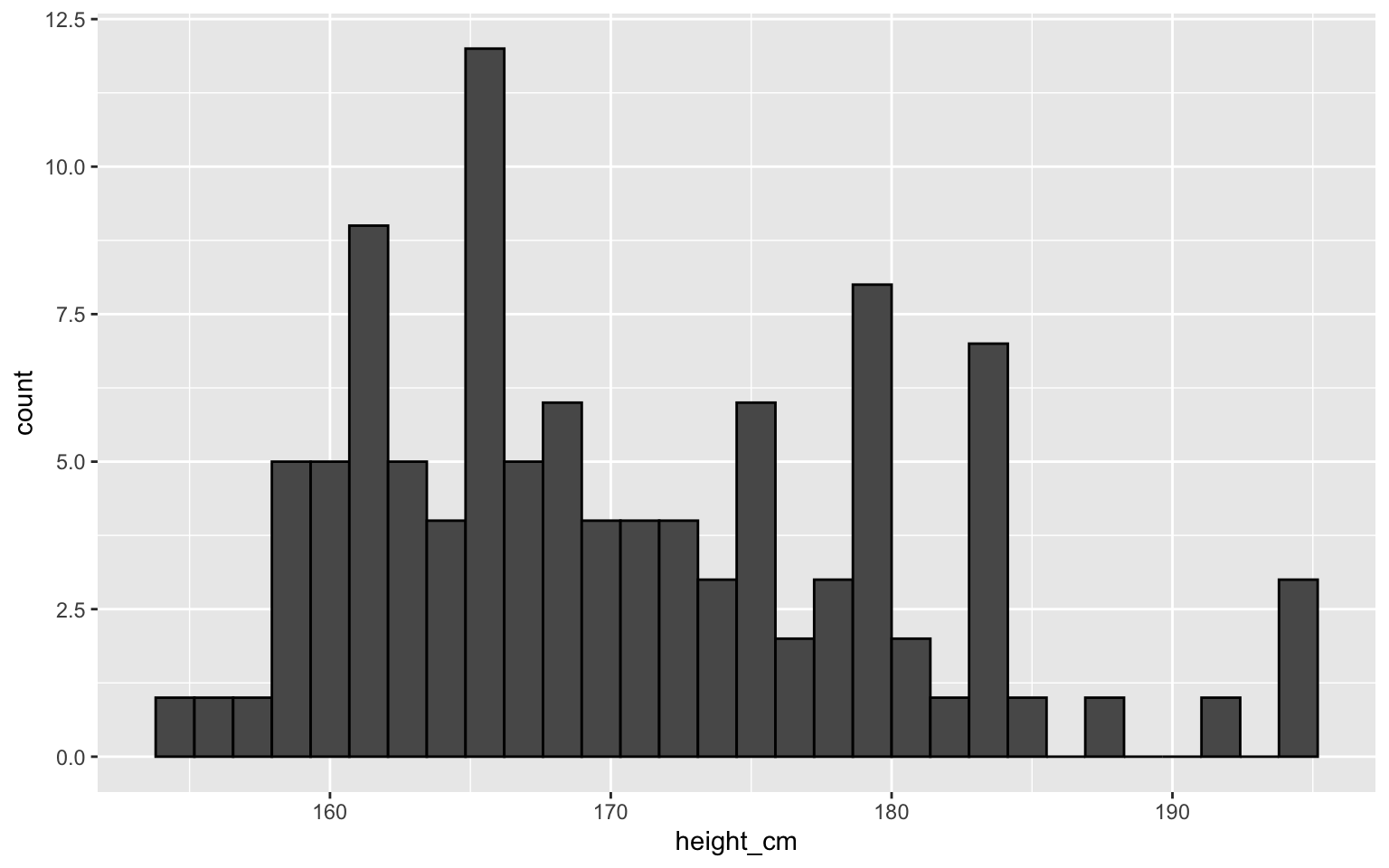
Histogram for numeric variables
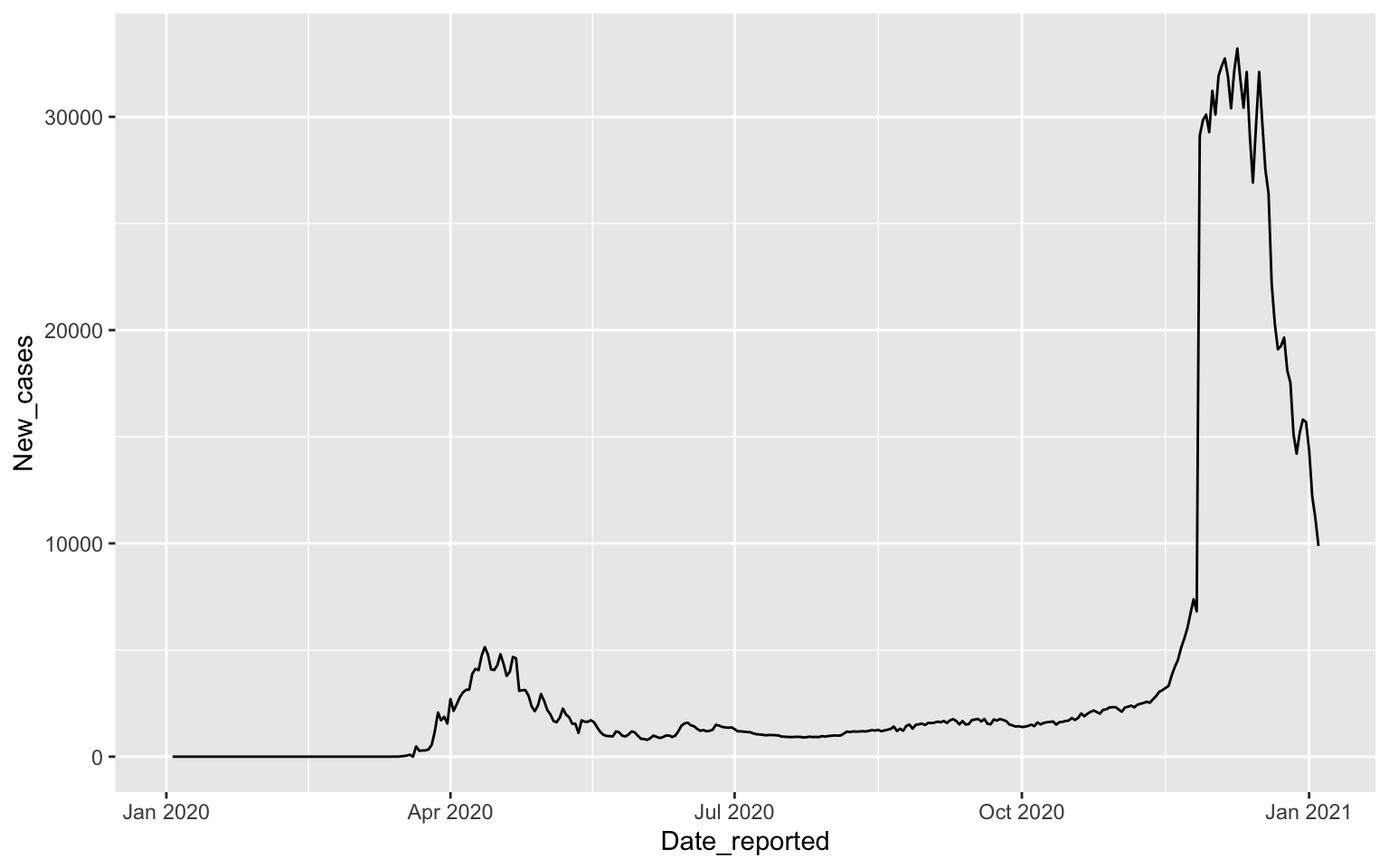
Line Plots
COVID-19 cases depend on time
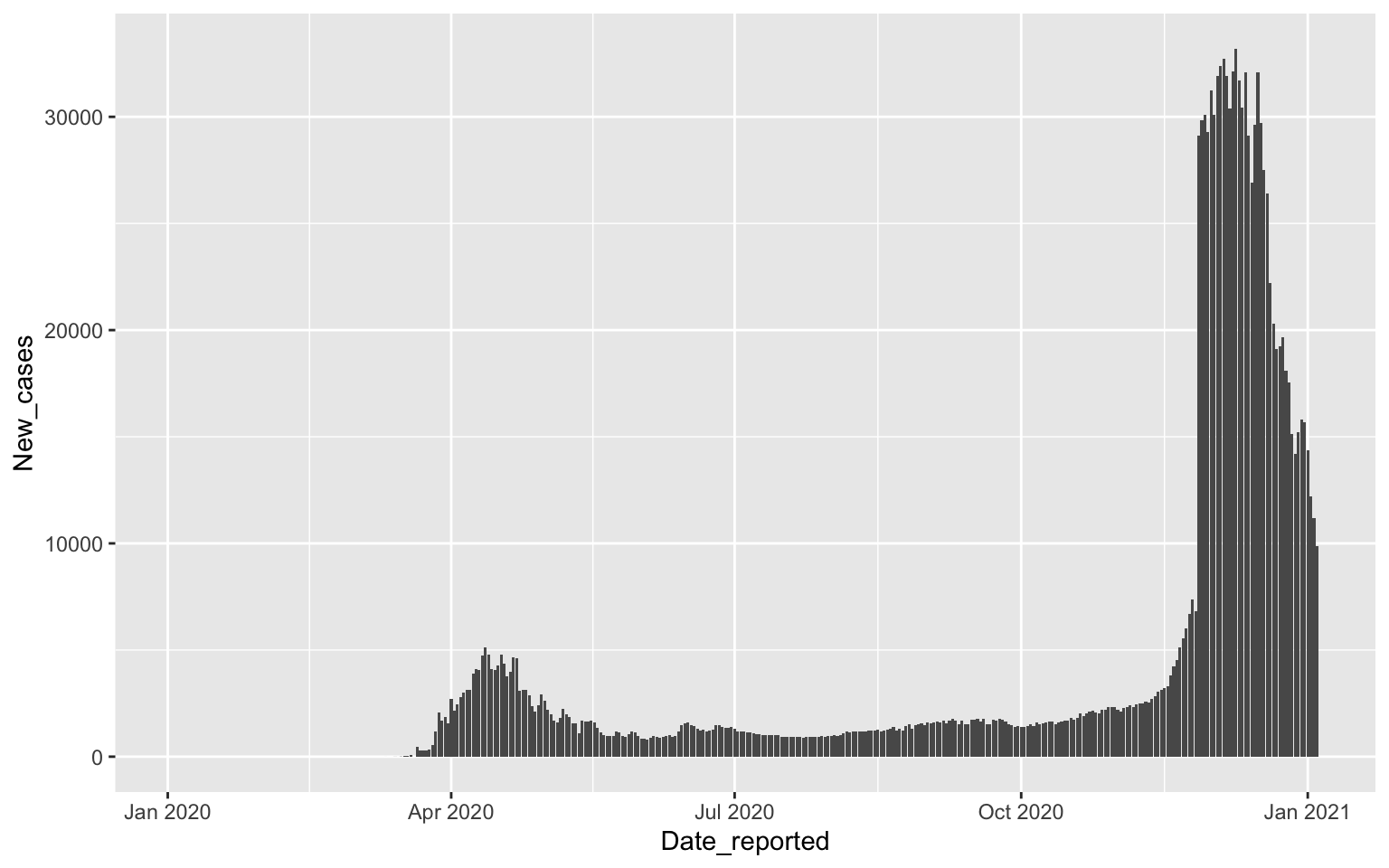
Drawing columns
Focusing on April
Turkey %>%
filter(Date_reported>"2020-03-31", Date_reported< "2020-05-01") %>%
ggplot(aes(x=Date_reported, y=New_cases)) + geom_col()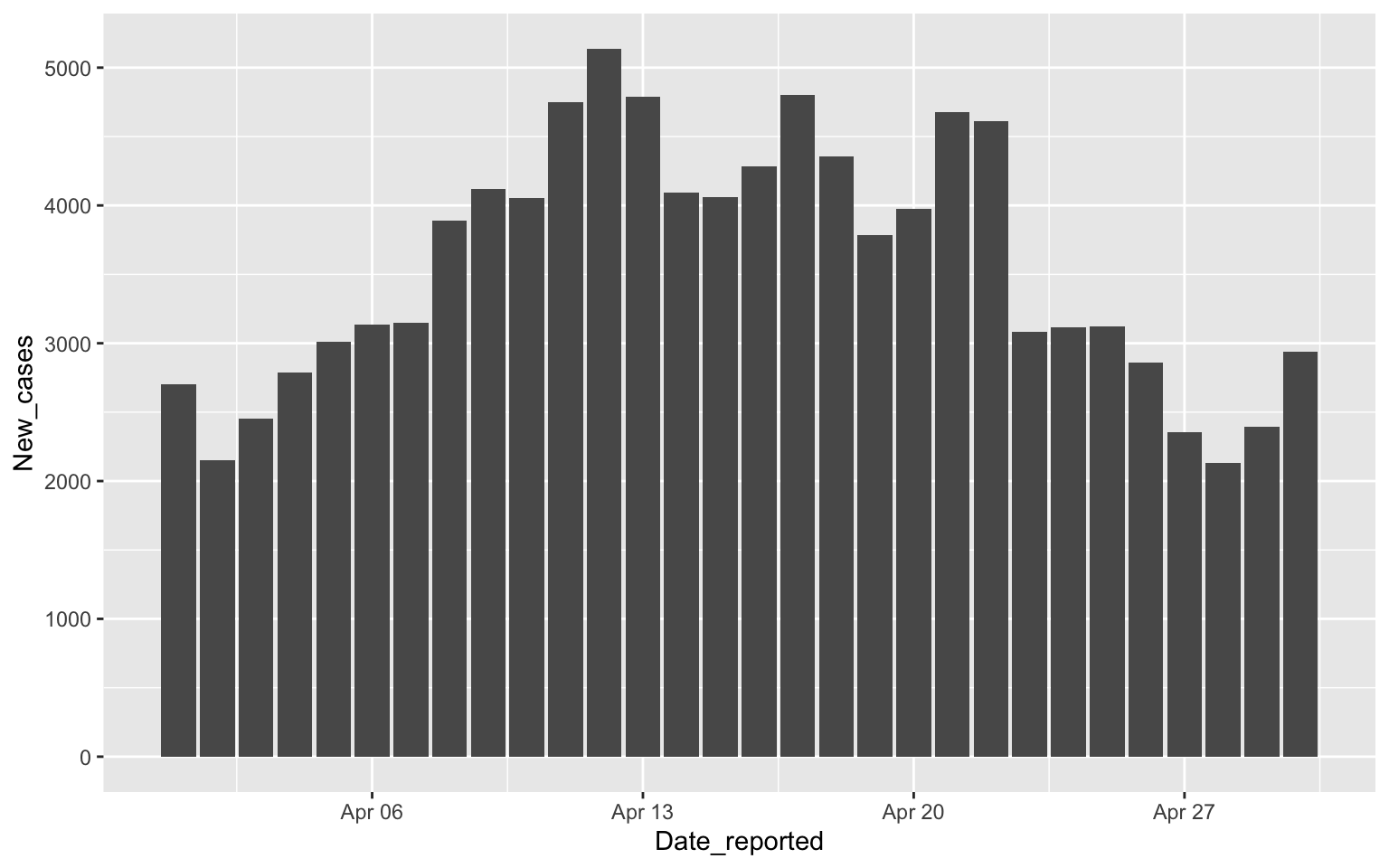
Drawing all European Countries
covid %>% filter(WHO_region=="EURO") %>%
ggplot(aes(x=Date_reported, y=Cumulative_cases, color=Country)) +
geom_line()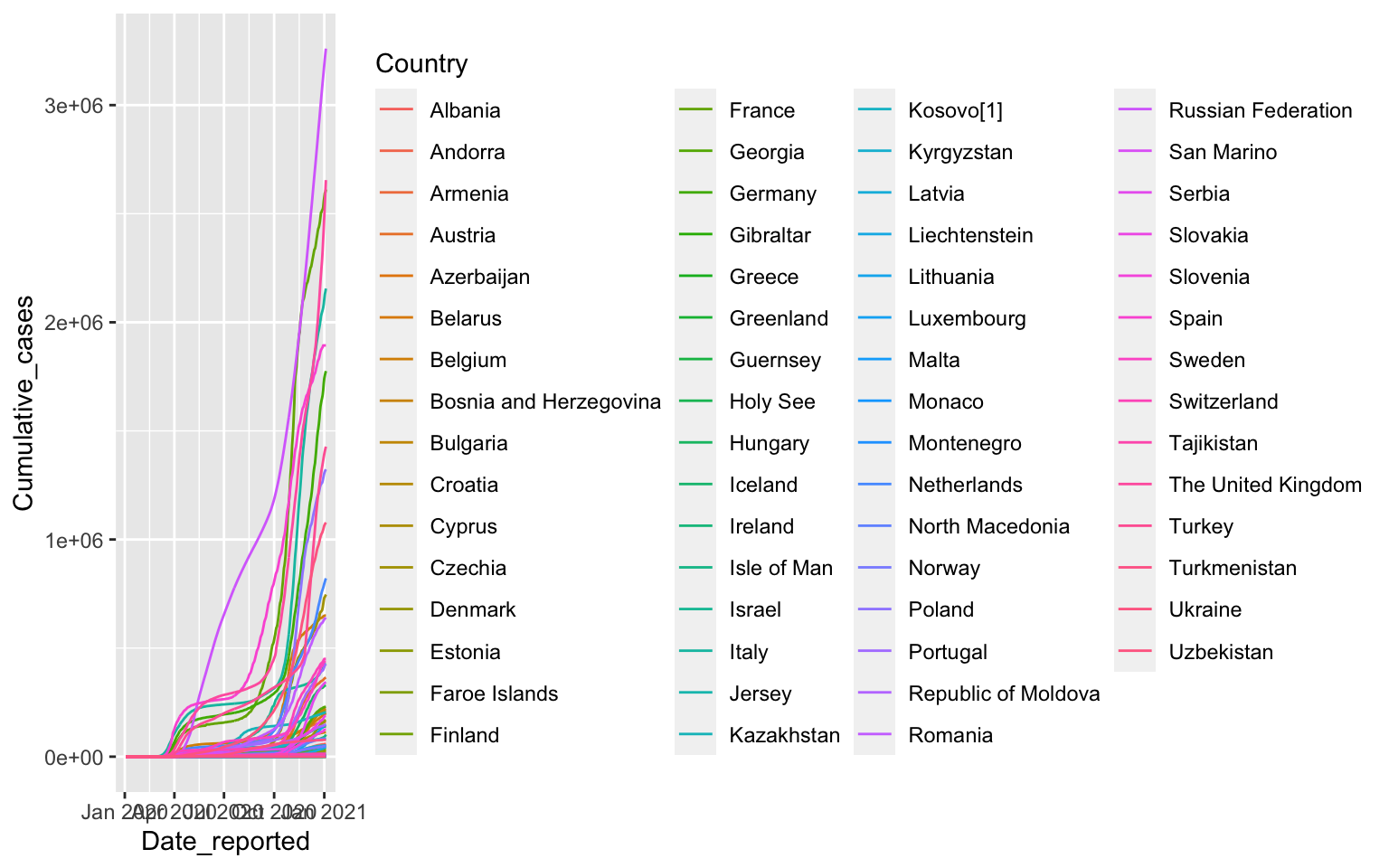
Which region has few countries?
# A tibble: 7 x 2
WHO_region `n_distinct(Country)`
<chr> <int>
1 AFRO 50
2 AMRO 56
3 EMRO 22
4 EURO 62
5 Other 1
6 SEARO 11
7 WPRO 35The easiest case is SEARO: South East Asia
South East Asia
covid %>%
filter(WHO_region=="SEARO") %>%
ggplot(aes(x=Date_reported, y=New_cases, color=Country)) +
geom_line()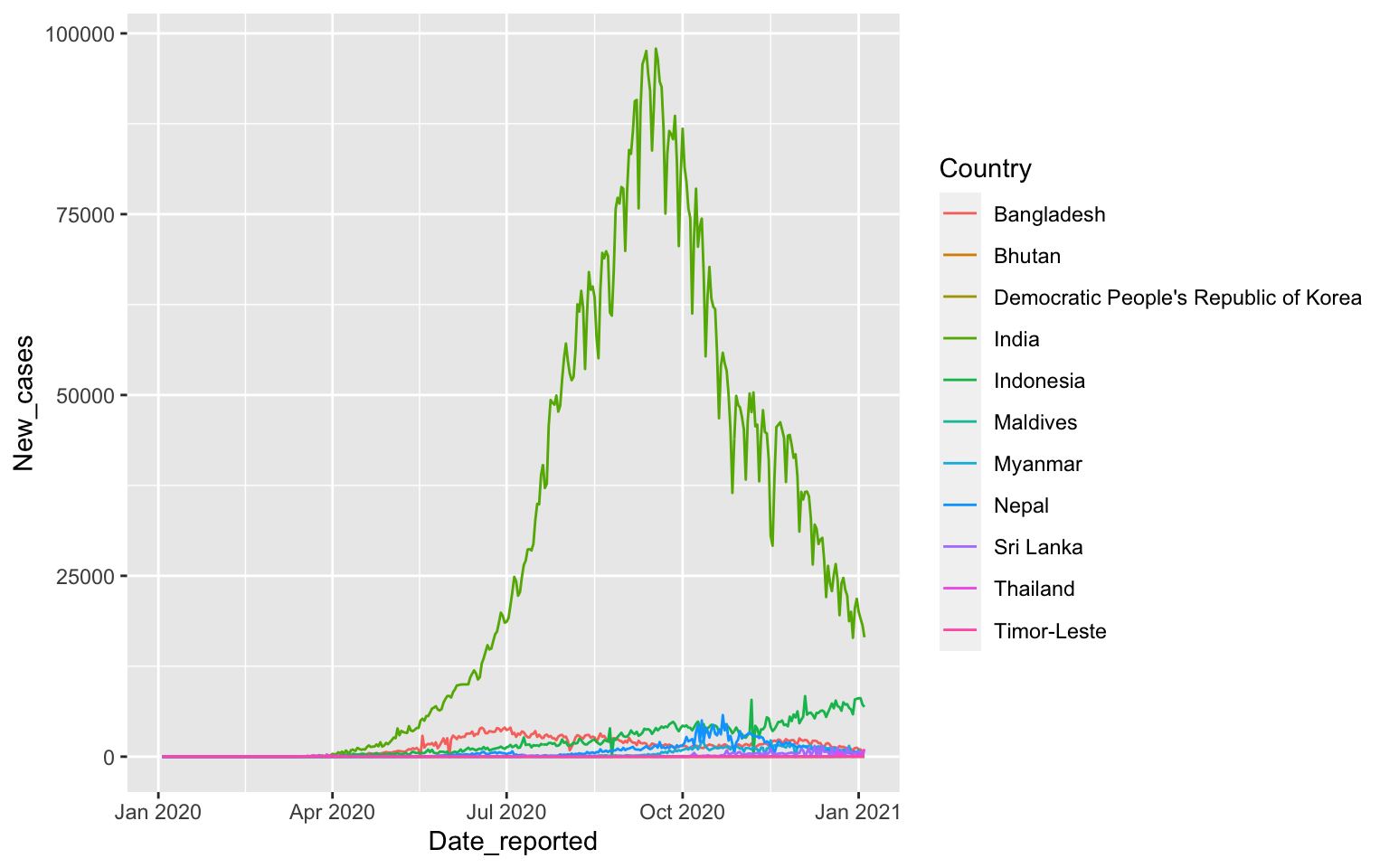
South East Asia in April
covid %>% filter(WHO_region=="SEARO") %>%
filter(Date_reported>"2020-03-31", Date_reported< "2020-05-01") %>%
ggplot(aes(x=Date_reported, y=New_cases, color=Country)) +
geom_line()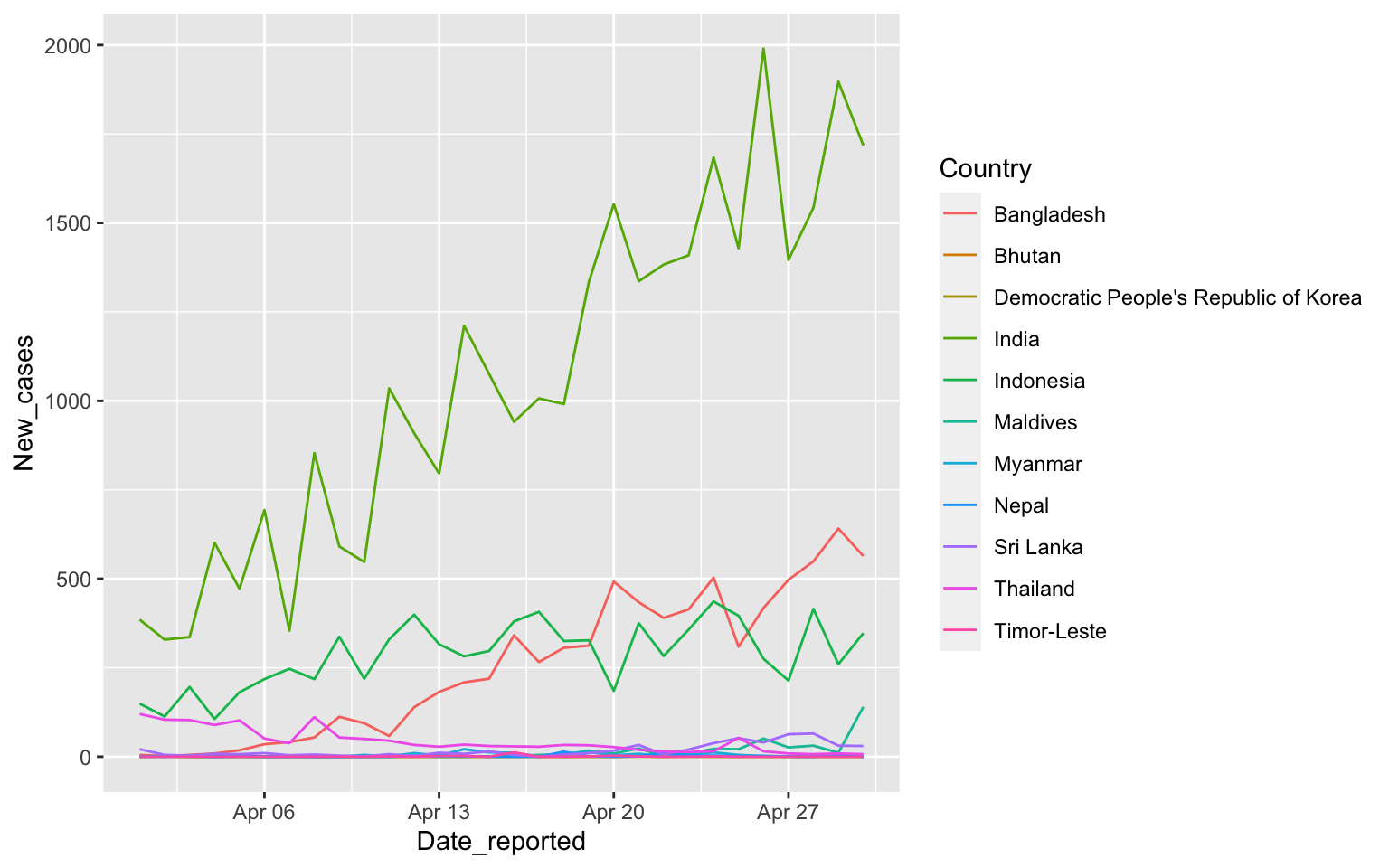
South East Asia with columns
covid %>% filter(WHO_region=="SEARO",
Date_reported>"2020-03-31", Date_reported<"2020-05-01") %>%
ggplot(aes(x=Date_reported, y=New_cases, color=Country)) +
geom_col()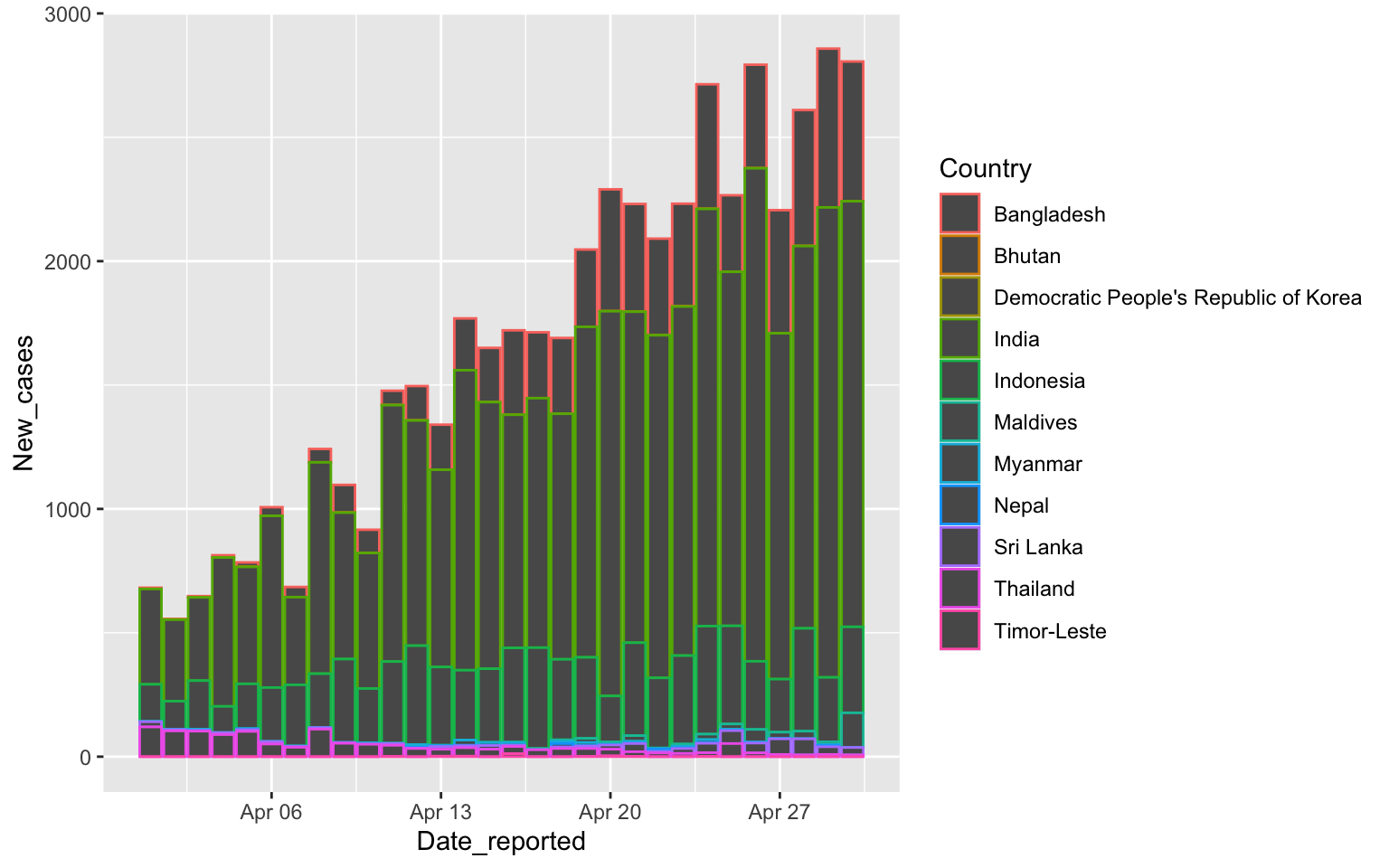
Columns have color and fill
covid %>% filter(WHO_region=="SEARO",
Date_reported>"2020-03-31", Date_reported< "2020-05-01") %>%
ggplot(aes(x=Date_reported, y=New_cases, fill=Country)) +
geom_col()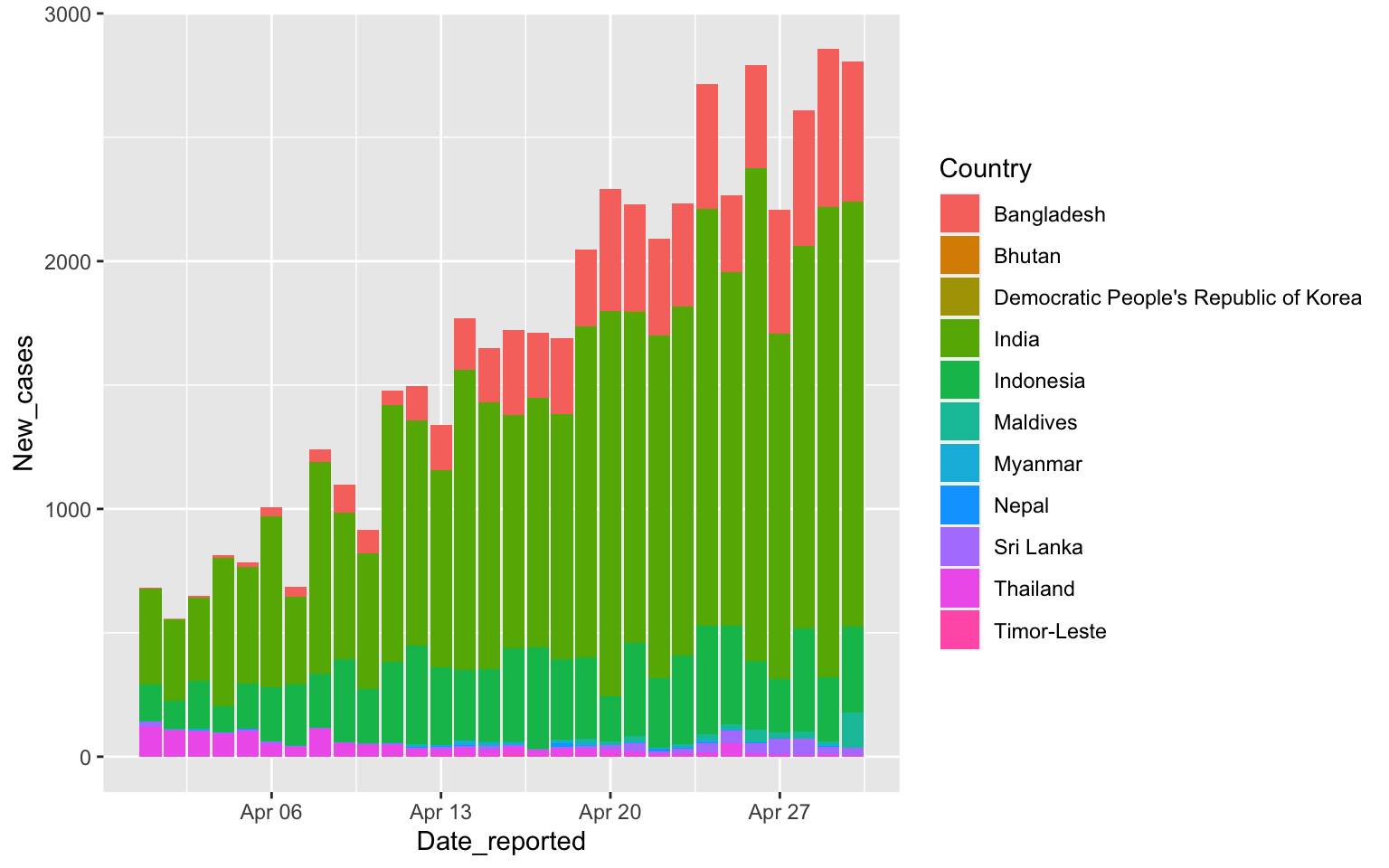
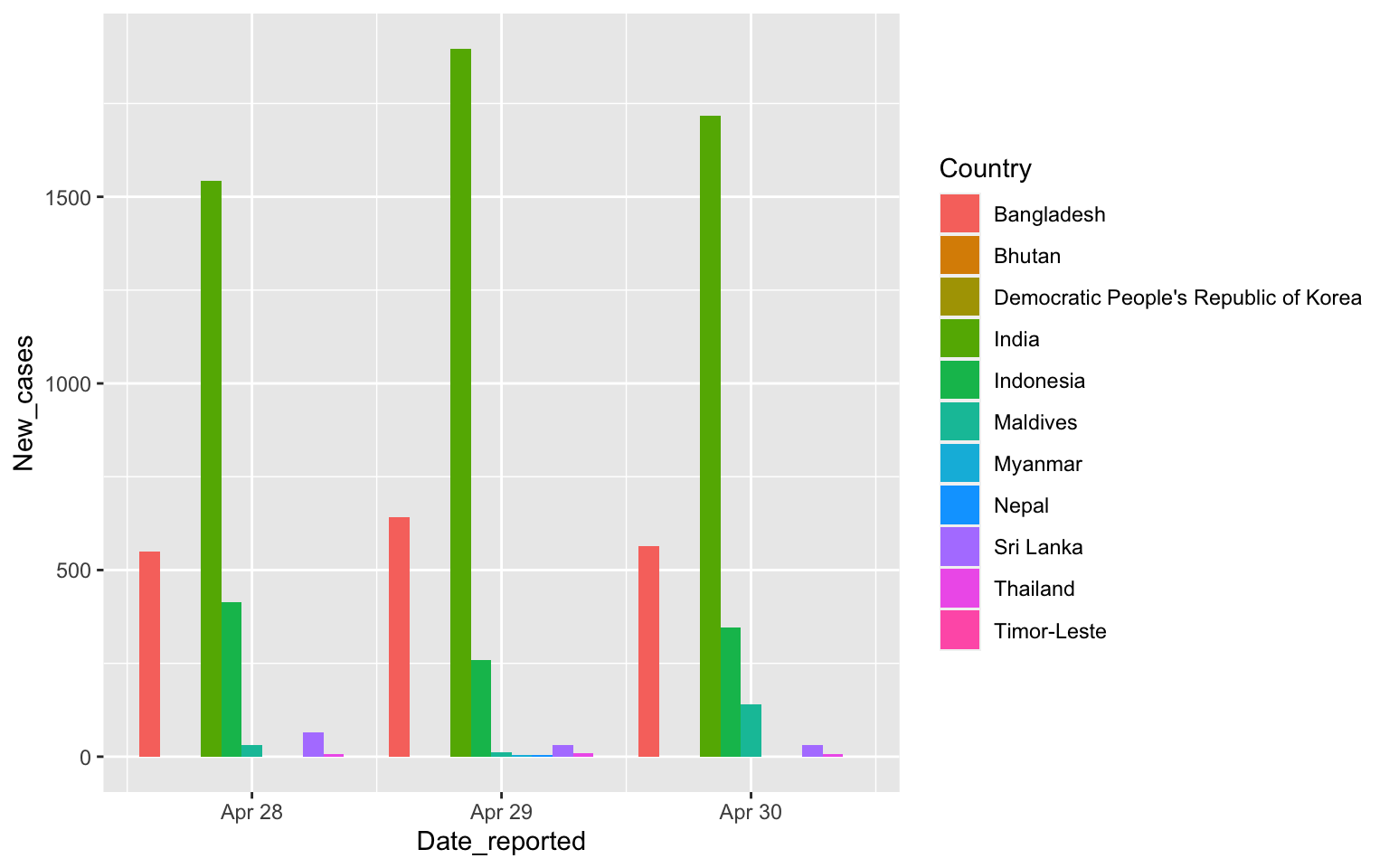
Putting Columns side to side
covid %>% filter(WHO_region=="SEARO",
Date_reported>"2020-04-27", Date_reported< "2020-05-01") %>%
ggplot(aes(x=Date_reported, y=New_cases, fill=Country)) +
geom_col(position=position_dodge())