Class 5: Searching and comparing sequences
Bioinformatics
Andrés Aravena
6 November 2020
How does spell check work?
When you write a text in any good text editor
(or even in Microsoft Word®),
some words are automatically underlined with red
These are words that are not found in the dictionary
Sometimes the editor can suggest the correct word
How does the computer do that?
What do you think?
There are two approaches
One option is to define a rule, like a program, that will tell if a word is correct or not \[ isCorrect: Words ↦ \{True, False\} \] That is hard, in general
Other option is to have a dictionary or database of know correct words, and a way to find the nearest word \[ Nearest: (Words, Database) ↦ \{Words\in Database\} \]
Searching sequences is the same problem
Searching a sequence
Let’s say we have a sequence and we want to identify it
Our sequence is called query
We look for it on a list of known sequences
The list is called database
Each sequence is a subject
Database
For this example, please download a sample database
Fasta format
http://www.dry-lab.org/static/bioinfo/short-protein.faaComma-separated values
http://www.dry-lab.org/static/bioinfo/short-protein.csv
How can you see the content of these files in your computer?
Lets search some sequences
query 1: ASELLKYLTT
query 2: ASELLKALTT
query 3: ASELLKYALTT
query 4: ASELLKLTTDo the search in your computer
What did you find?
How to fix a failed search
If you did it correctly, only the first search gave any result
The rest did not find anything
But we can see that the queries are very similar
How can we find sequences that are similar but not identical?
What do we mean by similar?
When are two sequences similar?
We can calculate a distance between strings
First idea:
We count the number of mismatches
Both strings must have the same length
ASELLKYLTT ASELLKALTT
Here distance(ASELLKYLTT,ASELLKALTT) is 1
This is called Hamming distance
Hamming distance examples
How many substitutions we need to go from one sequence to the other
CATandCAThave a Hamming distance of 0CATandBAThave a Hamming distance of 1CATandBAGhave a Hamming distance of 2
How does database search work
We look for the smallest distance
We calculate the distance between our query and each subject in the database
If the distance is 0, we have found a perfect match
Let’s make a Hamming distance calculator in Excel or Google Sheets
Let’s pause a minute and do this exercise
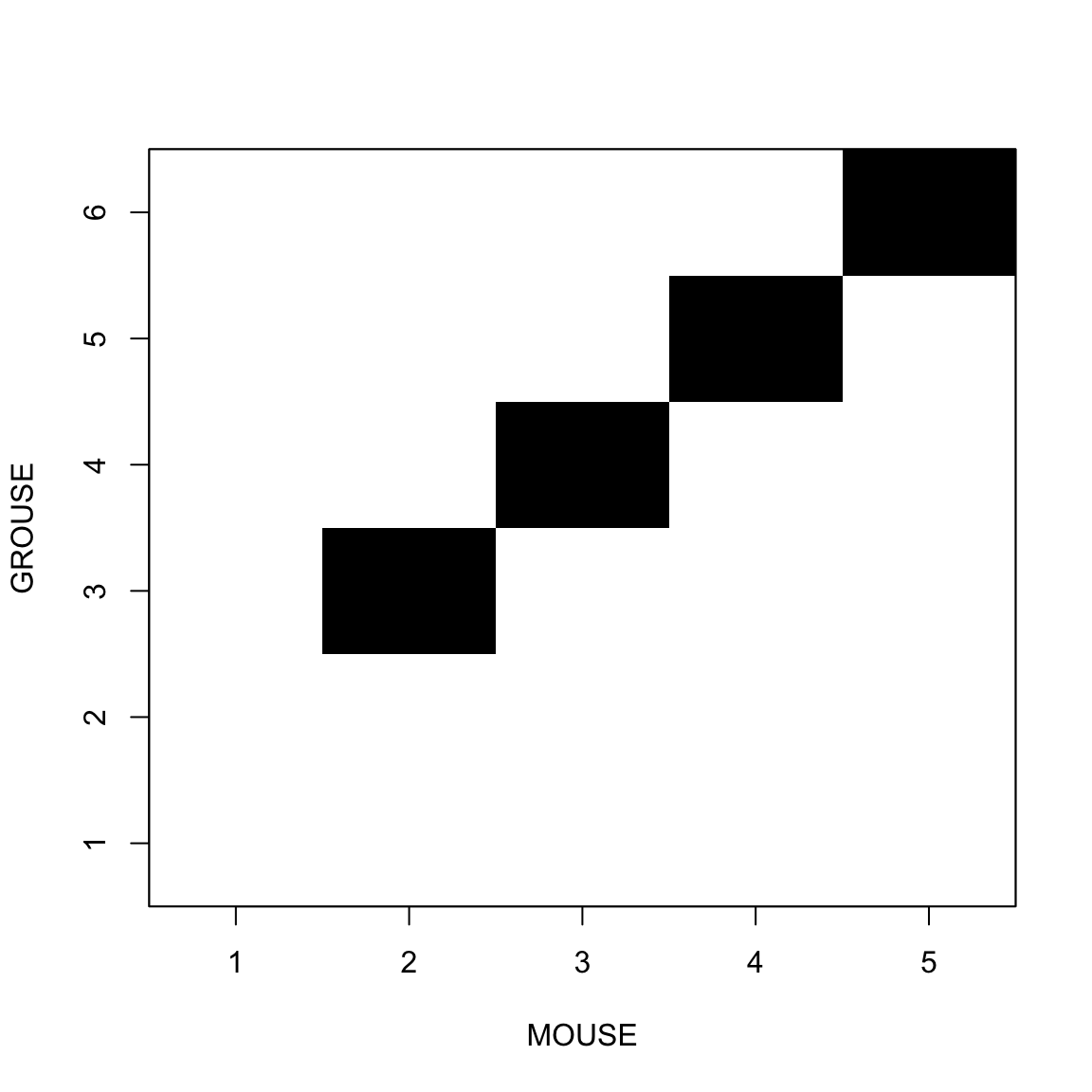
When sequences have different length
In that case we need to insert “gaps”
MOUSE-
GROUSEHamming Distance=6
Gaps are represented by -
Placing two sequences face to face, inserting gaps if necessary is called Pairwise Alignment
There are many ways to insert gaps
The distance depends on the gaps positions
-MOUSE
GROUSEHamming Distance=2
MOUSE--
-GROUSEHamming Distance=7
So, which one is the distance?
In geometry we say that “the distance between a point X and a line L is the shortest length from X to any point in L”
Same idea
If there many “candidate distances”, we choose the smallest one
This is an important idea
Distance is the length of the shortest path
Now we also count gaps
Hamming distance counts substitutions between sequences
Now we counts substitutions, insertions and deletions
This is called Levenstein distance
Hamming versus Levenstein
Hamming distance counts substitutions
ABCDEFGHIJ
BCDEFGHIJAHamming distance=10
Levenstein counts substitutions, insertions and deletions
ABCDEFGHIJK-
-BCDEFGHIJKALevenstein distance=2
What is the best way to insert gaps
If the sequence has m letters, there are 2m+1 ways to insert a single gap
CAT
CAT-
CA-T
CA-T-
C-AT
C-AT-
C-A-T
C-A-T--CAT
-CAT-
-CA-T
-CA-T-
-C-AT
-C-AT-
-C-A-T
-C-A-T-And that is not even counting larger gaps
We could also do
C--A-----TThere is a better way to find the best
We will see more details later
The idea is to draw a rectangle
- Query in the rows
- Subject in the columns
- Mark cells where row letter equals column letter
It looks like this
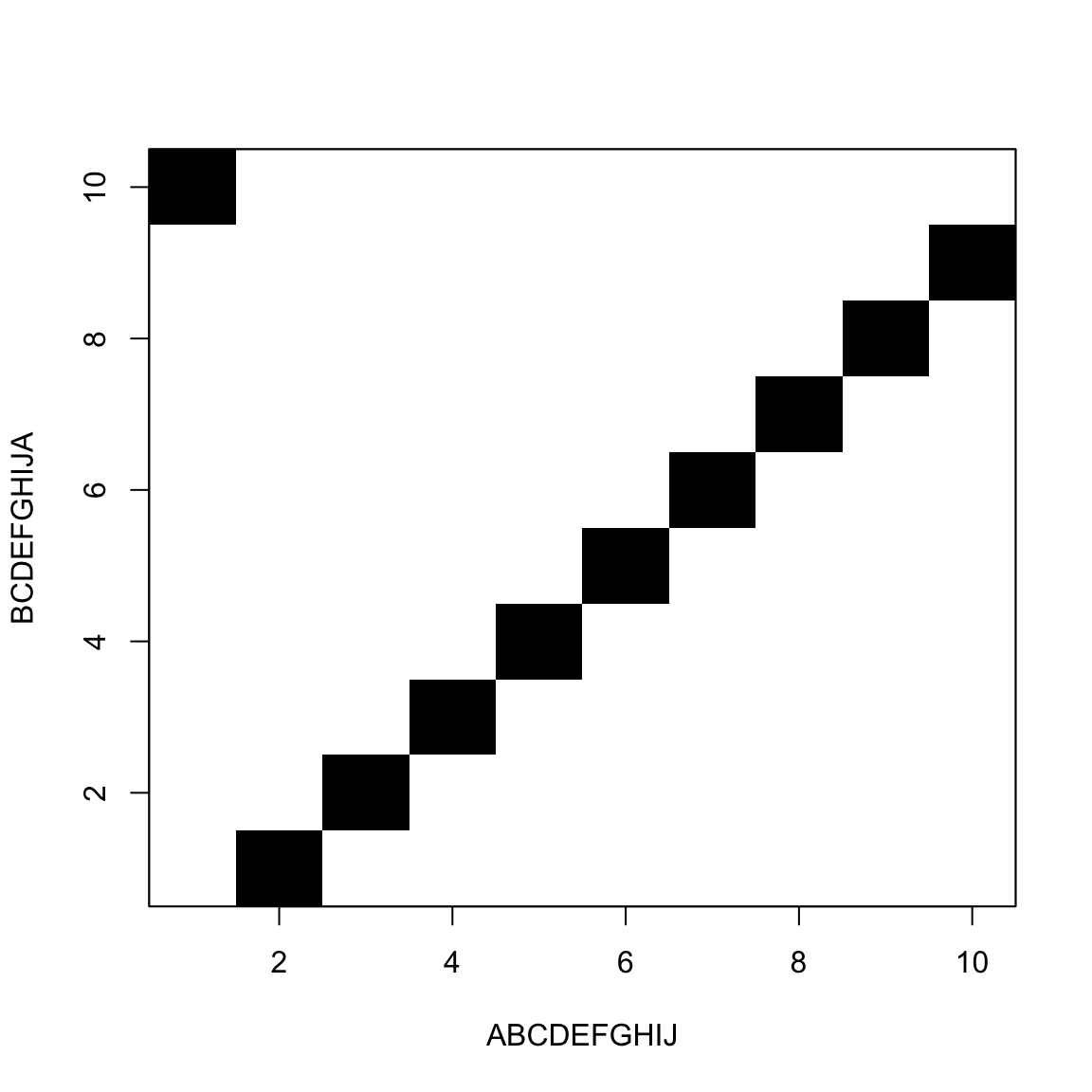
The goal is to move from one corner to the other
Jumping black to black is free
Horizontal and vertical moves are gaps
Exercise
Prepare a DotPlot in Excel or Google Sheets {.center background=“var(–good-blue)” .large .white}
We move from one corner to the other
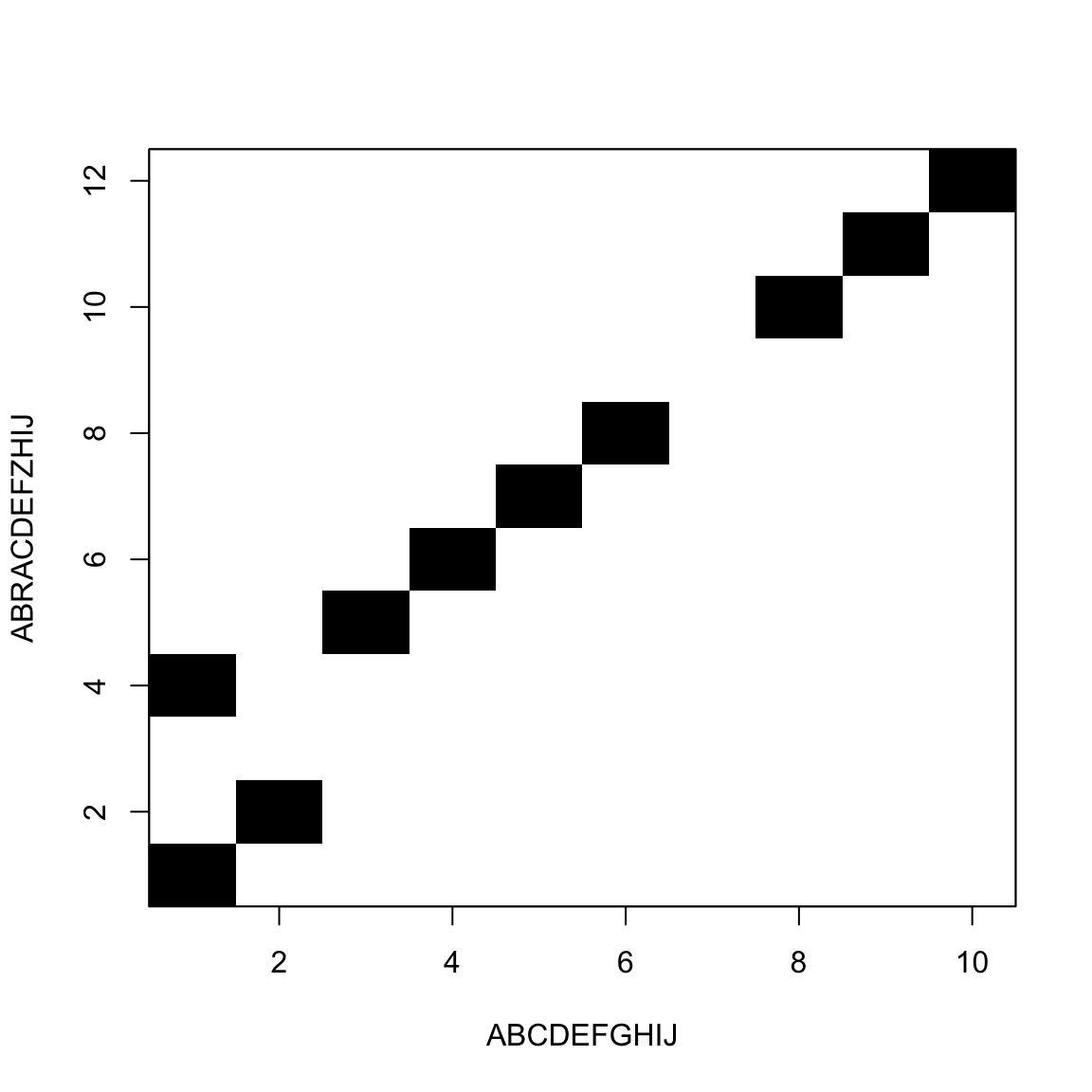
White blocks add to the distance
Comparing a sequence with itself
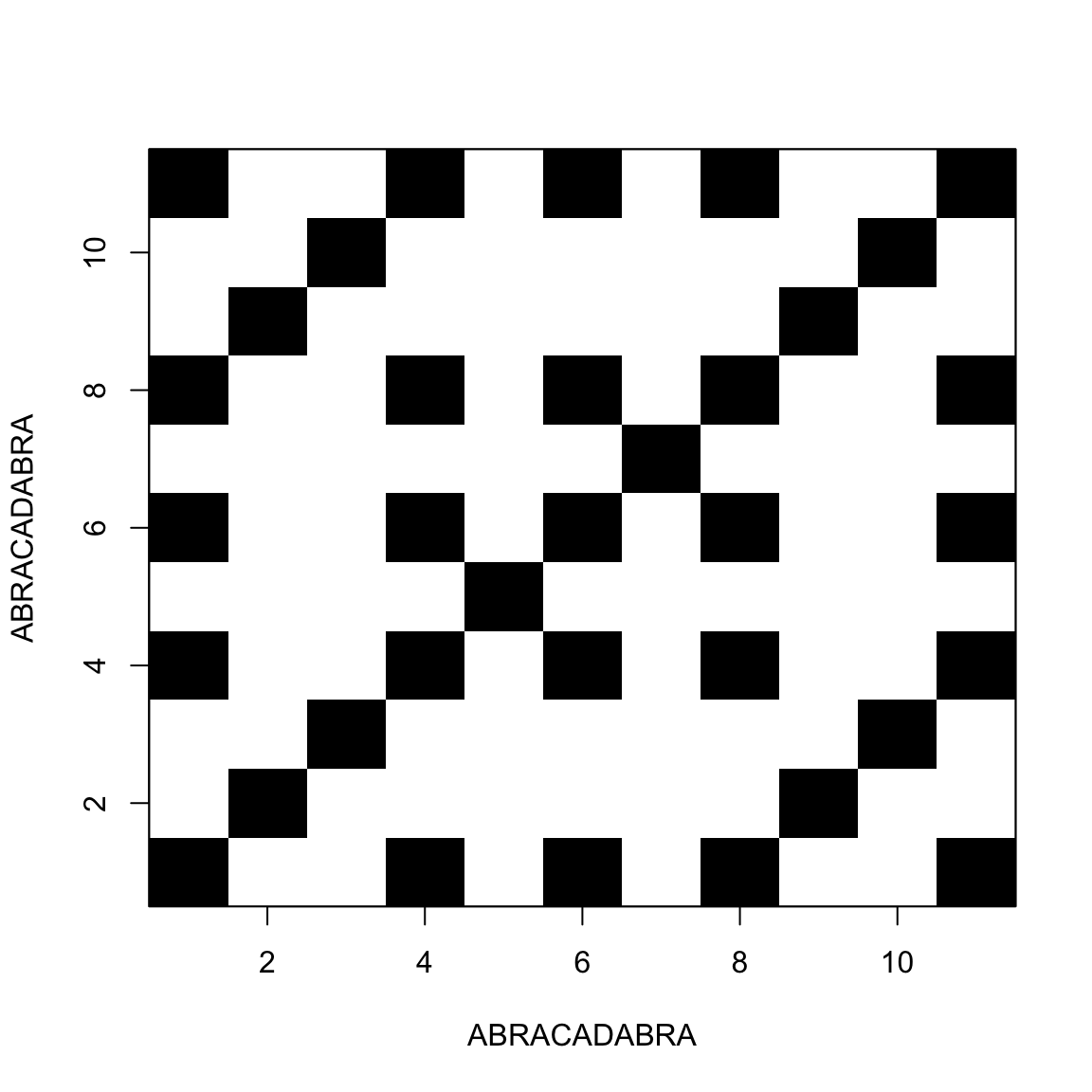
Comparing a sequence with itself
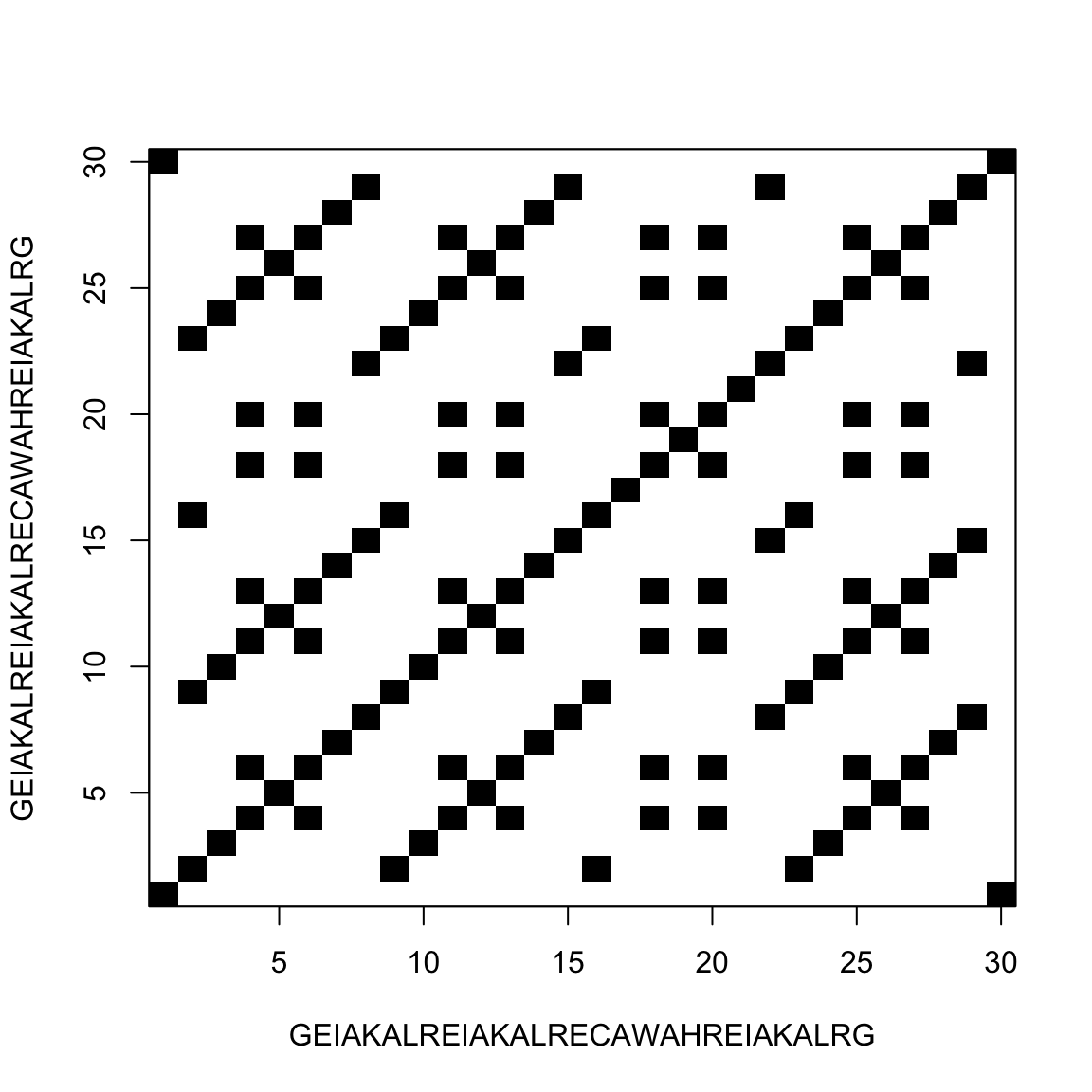 This allow us to find repeats inside the sequence
This allow us to find repeats inside the sequence
Palindromic sequences / Hairpins
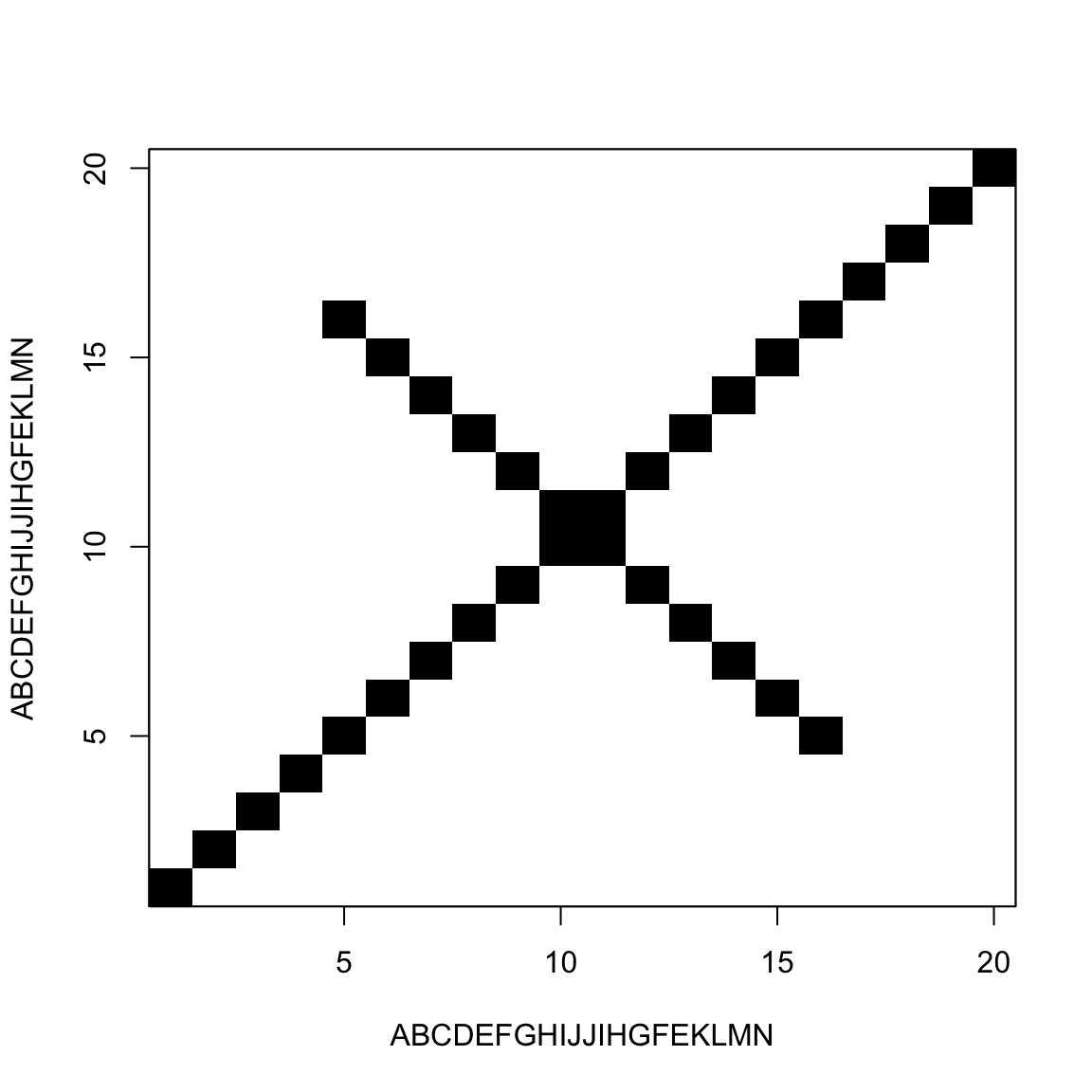 A sequence is said palindromic when it repeats itself backwards
A sequence is said palindromic when it repeats itself backwards
In RNA that results in a single-strand structure called hairpin
They are usually transcription terminators
Partial matching
Levenstein distance is Global Alignment
Useful to compare proteins
But not good when we look for a gene in a genome
or a domain in a protein
What shall we do when the query is much smaller than all subjects?
Semi-global alignment

In this case we want to go from one side to the other
Gaps in semi-global alignment
The query is much smaller than the subject
In this case we distinguish two kind of gaps
Internal gaps, inside the sequences
External gaps, outside the query sequence
External gaps do not count
External gaps are caused by the experiment
For example, we use PCR to cut part of a genome
Therefore anything outside the sequence cannot be seen for technical reasons
Internal gaps do count
Internal gaps are caused by nature
They are gained or lost during replication
They have biological meaning
Therefore, they must be counted
Summary
- Hamming distance
- Levenstein distance
- Dot plot
- Global alignment
- Semi-global alignment