Class 3: NCBI Entrez
Bioinformatics
Andrés Aravena
30 October 2020
Today’s Topics
Searching into NCBI
Advanced queries
Automatic queries
Taxonomy
Queries
NCBI Entrez queries
Searching NCBI has much more options than Google
(do you know Google options?)
By default the query text is searched in any part of any database
But you can specify the fields where you are looking for
- Title of a paper
- author
- date
- taxonomic id
Entrez Examples
protease NOT hiv1[organism]- This will limit the search to all proteases, except those in HIV 1.
1000:2000[slen]- This limits the search to entries with lengths between 1000 to 2000 bases for nucleotide entries, or 1000 to 2000 residues for protein entries.
Entrez Examples
10000:100000[mlwt]- This limits the search to protein sequences with calculated molecular weight between 10 kD to 100 kD.
src specimen voucher[properties]- This limits the search to entries that are annotated with a /specimen_voucher qualifier on the source feature.
Creating advanced queries
Quotes " are important
The fields are written inside brackets []
Each database page includes an Advanced Search option
Combining queries
Entrez queries can be single words, short phrases, sentences, database identifiers, gene symbols, or names
AND: Finds documents that contain terms on both sides of the operator terms. The intersection of both searches.
OR: Finds documents that contain either term. The union of both searches.
NOT: Finds documents that contain the term on the left but not the term on the right of the operator. The subtraction of the right side from the left side
Example
AND must be in uppercase. It is recommended to also use uppercase for OR and NOT
Operators are processed left-to-right
promoters OR response elements NOT human AND mammalsParenthesis can be used to control the evaluation order
g1p3 AND (response element OR promoter)
Dates and Other Ranges
Certain fields can accept ranges of values
- Publication Date, Modification Date, Accession, Molecular Weight, and Sequence Length
Low and high numbers are entered with a colon “:” between them followed by the field
110:500[Sequence Length] 2015/3/1:2016/4/30[Publication Date]
NCBI online documentation
We can get a different explanation in the public documentation made by NCBI
All documents made by NCBI are public domain
Automatization
Pipelines: putting all together
When we design molecular biology experiments, or when we analyze their results, we need to use several tools in chain
Today we are going to see an example using the NCBI website
F-Box protein domain

Our challenge is to find proteins in legumes having F-Box and WD-40 domains
Protein domains according to http://pfam.xfam.org/
We need an official definition of each domain
F-box
- motif PF00646.
- alternative motifs (Gupta et al. 2015)
- PF12937, PF13013, PF04300, PF07734, PF07735, PF08268 and PF08387
WD-40
- motif PF00400
Finding proteins with those domains
Using NCBI Conserved Domain Architecture Retrieval Tool (https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi)
Use the query:
[pfam00646,pfam00400]to look for proteins that contain both domains in the specified order
Results
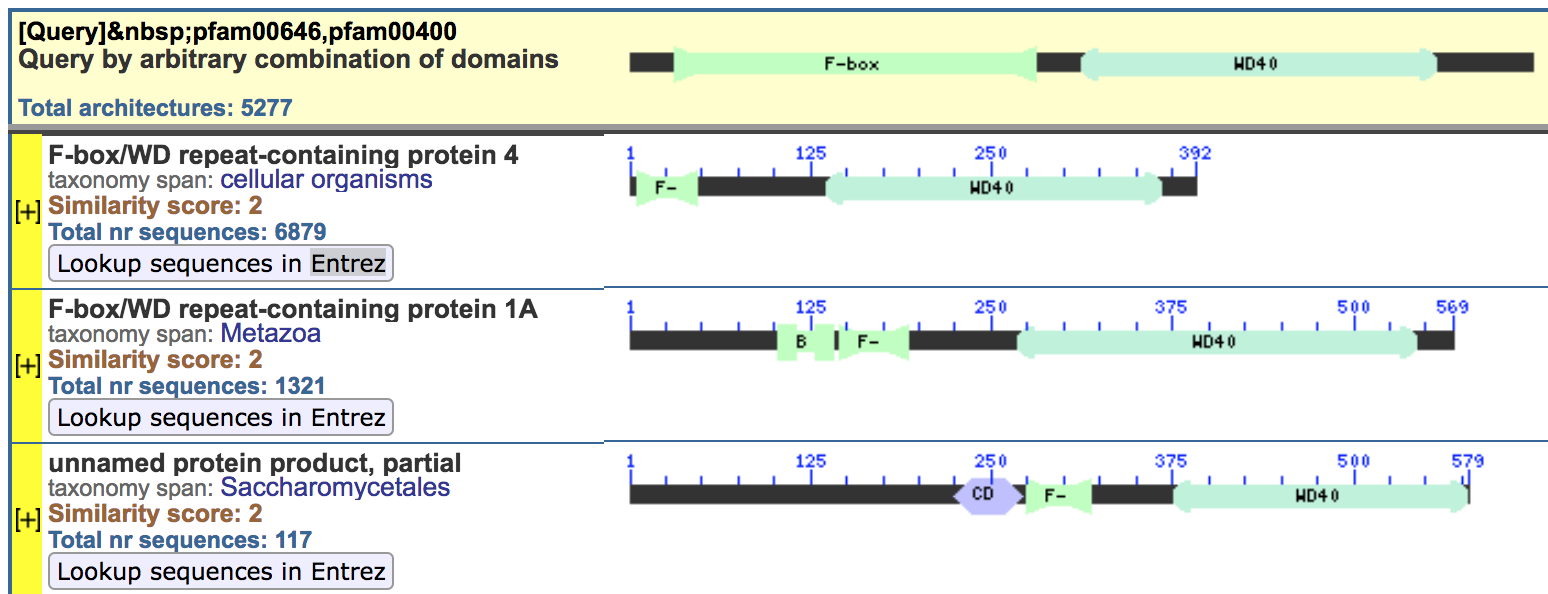
Results
- Several architectures contain both motifs
- Selected the first, with only the relevant domains and nothing else
- Click the button “Lookup sequences in Entrez” to find the list of proteins
Filter results: only Legumes
Most of times is a good idea to check the Taxonomy database
Each sequence on GenBank is tagged with a taxon id
Using taxid is more precise than using common names
For example, a protein from human can be labeled “95% similar to mouse”
Is that a human or a mouse protein?
Downloading
For your convenience you can download the sequences
- Decide Format
- Decide Content
In this case we only need accession ids
Finding more proteins
Now we use BLAST to find other proteins in legumes similar to the ones we have
Notice that CDART only has some proteins pre-processed. New sequences take time to be processed
How would you do that?
Save your search strategy
It is essential that your protocol can be replicated
It is a very good idea to save the search strategy in a file
It is also wise to save the output in a text file
Separate by tab or by comma
Process the output|check domains
How many new proteins we find?
Do all of them have the good domains?
Let’s use CDD again, this time looking for motifs on the new proteins \[protein \to\{domains\}\] (the first time was \(domain\to\{proteins\}\))
Next steps
- Keep only proteins with the good domains
- Download the sequences of all proteins
- Download the sequences of the messengers
- Design primers to measure gene expression
- Find literature about gene expression
It is boring to do it one by one
And takes a lot of time
It is easy to make mistakes
It is hard to replicate
Can we do it automatically?
E-tools: Entrez Pipelines
ESearch -> ESummary;
ESearch -> EFetch;
EPost -> ESummary;
EPost -> EFetch;
ESearch -> ELink;
EPost -> ELink;
EPost -> ESearch;
ELink -> ESearch;
ESearch -> ELink -> ESummary;
ESearch -> ELink -> EFetch;
EPost -> ESearch -> ESummary;
EPost -> ESearch -> EFetch;
EPost -> ELink -> ESearch -> ESummary;
EPost -> ELink -> ESearch -> EFetch;Map of E-tools
Use your favorite language
There are Entrez libraries for most languages
For example in R it is called rentrez
There is a command line version, and versions for all major computer languages