Class 1: Why do we care about Bioinformatics?
Bioinformatics
Andrés Aravena
23 October 2020
Welcome to “Bioinformatics”
Today’s ideas
- What “Bioinformatics” is and is not
- Why you should care
- How to get bioinformatic data for free
- What kind of data we can get
- What is important in the data
Bioinformatics
what it is and what it isn’t
Molecular Biology 101
- DNA
- RNA
- Proteins
- Metabolism
What is Bioinformatics?
- Genomics
- sequences of DNA, RNA, AA
- Transcriptomics
- gene’s expression
- Proteomics
- 3D structure and interactions
- Metabolomics
- metabolites
What Bioinformatics is not?
- Using computers in a hospital
- Handling patient information
- Laboratory Information Management
- Microscope image analysis
Big picture
for this course
Genomics
- DNA sequencing
- Pairwise Alignment
- Multiple Alignment
- Genome Assembly
- Primer design
- Finding Binding Sites
Transcriptomics
Measuring gene expression
- qPCR
- Microarrays
- RNAseq
Mostly about statistics
Proteomics
- Find protein sequence
- mass spectrometry
- Find protein structures
- X-ray diffraction analysis
- Computational Biology prediction
- Find protein-protein interactions
What we should do here
- Role
- Concepts
- Statistics
- Access
- Tools
- Pathways
- Metagenomics
- Scripting
- Software
- Computational environment
Sayres, et al. “Bioinformatics Core Competencies for Undergraduate Life Sciences Education.”
PLoS ONE 13, no. 6 (2018): 1–20. https://doi.org/10.1371/journal.pone.0196878.
Detail
- Role: Understand the role of computation and data mining in hypothesis-driven processes within the life sciences
- Concepts: Understand computational concepts used in bioinformatics, e.g., meaning of algorithm, bioinformatics file formats
- Statistics: Know statistical concepts used in bioinformatics, e.g., E-value, z-scores, t test, type-1 error, type-2 error, employ R
Detail
- Access genomic: Know how to access genomic data, e.g., in NCBI nucleotide databases
- Tools genomic: Be able to use bioinformatics tools to analyze genomic data, e.g., BLASTN, genome browser
- Access expression: Know how to access gene expression data, e.g., in UniGene, GEO, SRA
- Tools expression: Be able to use bioinformatics tools to analyze gene expression data, e.g., GeneSifter, David, ORF Finder
Detail
- Access proteomic: Know how to access proteomic data, e.g., in NCBI protein databases
- Tools proteomic: Be able to use bioinformatics tools to examine protein structure and function, e.g., BLASTP, Cn3D, PyMol
- Access metabolomic: Know how to access metabolomic and systems biology data, e.g., in the Human Metabolome Database
Detail
- Pathways: Be able to use bioinformatics tools to examine the flow of molecules within pathways/networks, e.g., Gene Ontology, KEGG
- Metagenomics: Be able to use bioinformatics tools to examine metagenomics data, e.g., MEGA, MUSCLE
- Scripting: Know how to write short computer programs as part of the scientific discovery process, e.g., write a script to analyze sequence data
Detail
- Software: Be able to use software packages to manipulate and analyze bioinformatics data, e.g., Geneious, Vector NTI Express, spreadsheets
- Computational environment: Operate in a variety of computational environments to manipulate and analyze bioinformatics data, e.g., Mac OS, Windows, web- or cloud-based, Unix/Linux command line
What we really do here
We focus on How to understand results
- Role: What is bioinformatics
- Access: using NCBI, EBI
- Concepts: file formats and more
- Tools: understanding tools output
- Statistics: E-values, error type-1 and type-2
Concepts
- Pairwise Alignment
- Global
- Semi-global
- Local
- Multiple Alignment
- Cost
- Heuristics
- Trees
- Taxonomy
- Phylogenetic
- Ontology
Any Questions?
Why you should care
about bioinformatics
Technology changes fast
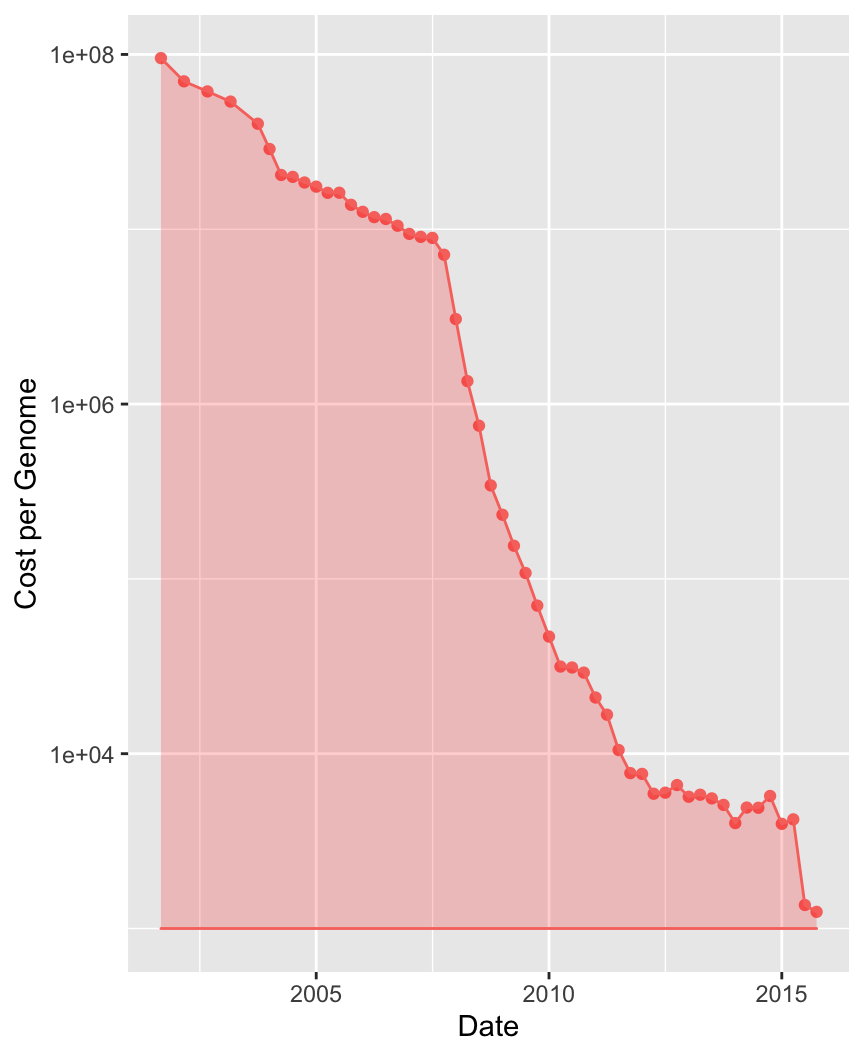
In 2001, the cost of sequencing the first human genome was USD 108
Today you can have your own genome for 1000 USD
The problem is no longer how to do the experiment
Instead is how do we make sense of the results
Manual jobs are now done by computers
Will a robot replace you?

Any Questions?
Four Paradigms of Science
According to Microsoft
1 Empiric
(since prehistoric times)

- observation of isolated facts
- description of related facts
- e.g. Botany, naming stars, Arab astronomers, Galileo, Tycho Brahe, Carl Linneaus
2. Theoretical
(Renaissance)

- Abstract models and theories
- Usually expressed in mathematical formulas
- Correct predictions validate the models
- e.g. Mendel laws of inheritance, Darwin natural selection theory, Kepler law of planet’s motion, Newton’s law of Gravity
3. Simulation Based
(Mid 20th century)
- Models that cannot be expressed in formulas
- Formulas that cannot be solved
- e.g. Protein structure prediction, three body problem, galaxy modeling
- Computational Astronomy, Computational Biology
4. Data Based
(21st century)

- Discovering patterns hidden in data
- Huge volumes of data
- Complex interactions
- e.g. Bioinformatics, Astroinformatics, Data mining
- Big Data, Machine Learning
