A chemical reaction combines hydrogen and oxygen to produce water
To make it simple we will assume that the reaction is this: \[2H+O\leftrightarrow H_2O\]
Our system has 3 items:
- Hydrogen
- Oxygen
- Water
March 29, 2019
A chemical reaction combines hydrogen and oxygen to produce water
To make it simple we will assume that the reaction is this: \[2H+O\leftrightarrow H_2O\]
Our system has 3 items:
We represent hydrogen by H, oxygen by O, and water by W.
Each vector has the number of atoms or molecules for different times
We will measure the number of atoms and molecules in moles
The initial quantities are:
There are two reactions at the same time:
from hydrogen and oxygen into water
(that is, left to right),
from water into hydrogen and oxygen
(that is, right to left).
According to our rules, we have two rates:
reaction1_ratereaction2_rateIn chemistry, these rates are usually written as
k1k2Shorter and cleaner for this class
k1 * H * H * O
k2 * W
Then the main part of the simulation is
for(i in 2:N) {
d_W[i] <- k1*H[i-1]*H[i-1]*O[i-1] - k2*W[i-1]
d_O[i] <- -k1*H[i-1]*H[i-1]*O[i-1] + k2*W[i-1]
d_H[i] <- -2*k1*H[i-1]*H[i-1]*O[i-1] + 2*k2*W[i-1]
W[i] <- W[i-1] + d_W[i]
O[i] <- O[i-1] + d_O[i]
H[i] <- H[i-1] + d_H[i]
}
Create a water_formation function.
Inputs are:
N: Number of steps in the simulationH_ini: initial amount of hydrogen, default 1O_ini: initial amount of oxygen, default 1W_ini: initial amount of water, default 0k1: reaction 1 rate, default 0.1k2: reaction 2 rate, default 0.1One atom of oxygen has 16 times the mass of one atom of hydrogen
Therefore one molecule of water has mass 18
Show that the total mass never changes
The Polymerase Chain Reaction (PCR) is a method used to synthesize millions of copies of a given DNA sequence.
A typical PCR reaction consists of series of cycles:
This loop is repeated between 25 and 30 times
We can represent the PCR reaction as a system with two parts, DNA and primers and one process, the thermal cycle.
Here we simplify the reaction and we forget about the polymerase and the dNTP. They both will be represented by primers.
The system is represented by this diagram:
Please write a function to simulate the PCR reaction.
The function name should be pcr and it must take four inputs:
N,dna_ini,primer_ini, andrate.The function must return a data frame with dna and primer.
If all is right, you should see this result
pcr(N=6, dna_ini=1e6)
dna primer 1 1000000 100000000 2 2000000 99000000 3 3980000 97020000 4 7841396 93158604 5 15146331 85853669 6 28150012 72849988
conc <- pcr(N=20, dna_ini=1e6) plot(conc$dna)
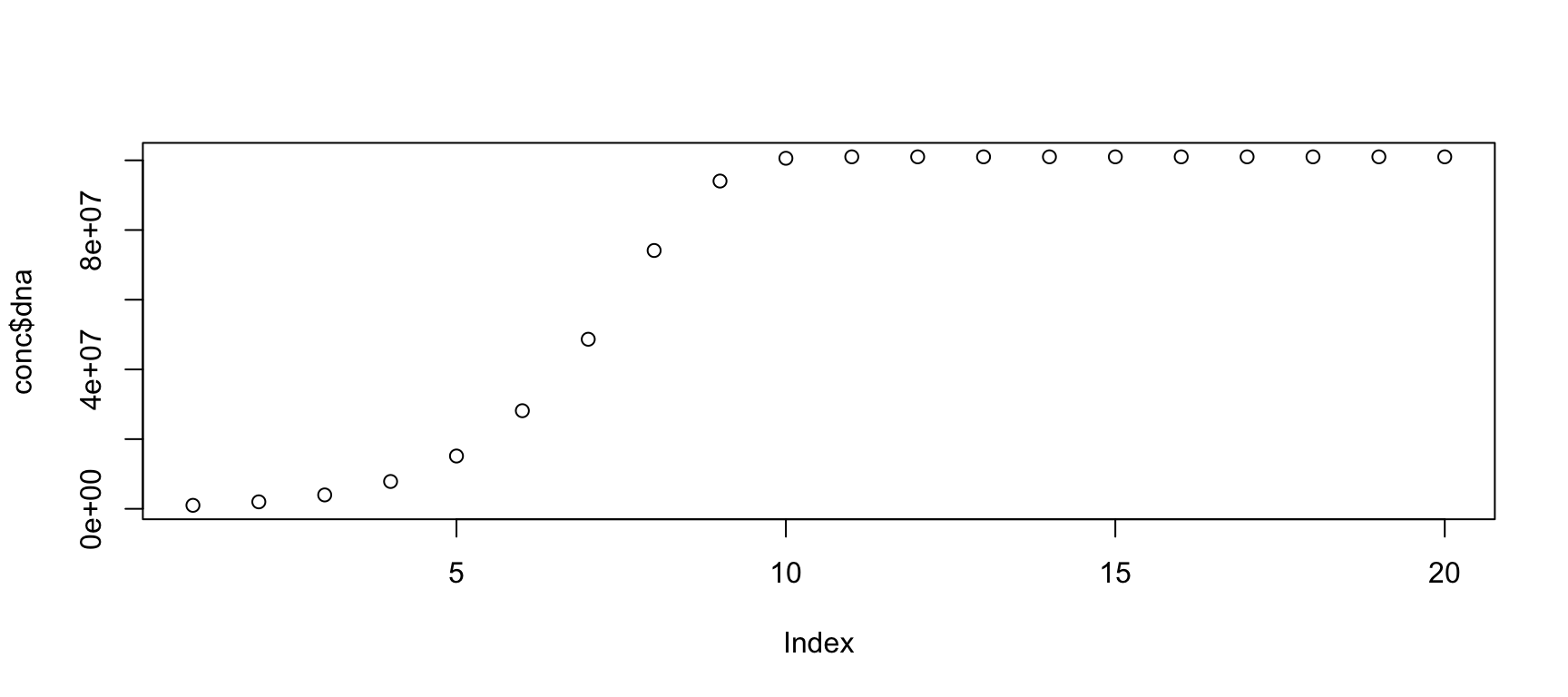
The PCR reaction curve depends on the initial concentration of DNA.
We want to understand this dependency for the following values of initial DNA concentration:
initial_dna <- 10**(0:6) initial_dna
[1] 1e+00 1e+01 1e+02 1e+03 1e+04 1e+05 1e+06
You have to create a data frame a list named conc.
This data frame has seven columns, with the result of pcr() for each of the initial DNA concentrations in initial_dna.
That is, from one DNA molecule per liter up to one million molecules per liter.
Simulate the PCR reaction for 30 cycles, and store only the value of the dna column.
If you did it correctly, you can use this code to create a plot.
plot(x=c(1,30), y=c(0, max(conc)), type="n",
xlab="PCR cycle", ylab="DNA Concentration",
main="Effect of initial DNA on PCR")
legend("topleft", legend=initial_dna, pch=1:7)
for(i in 1:ncol(conc)) {
points(conc[[i]], pch=i, type="b")
}
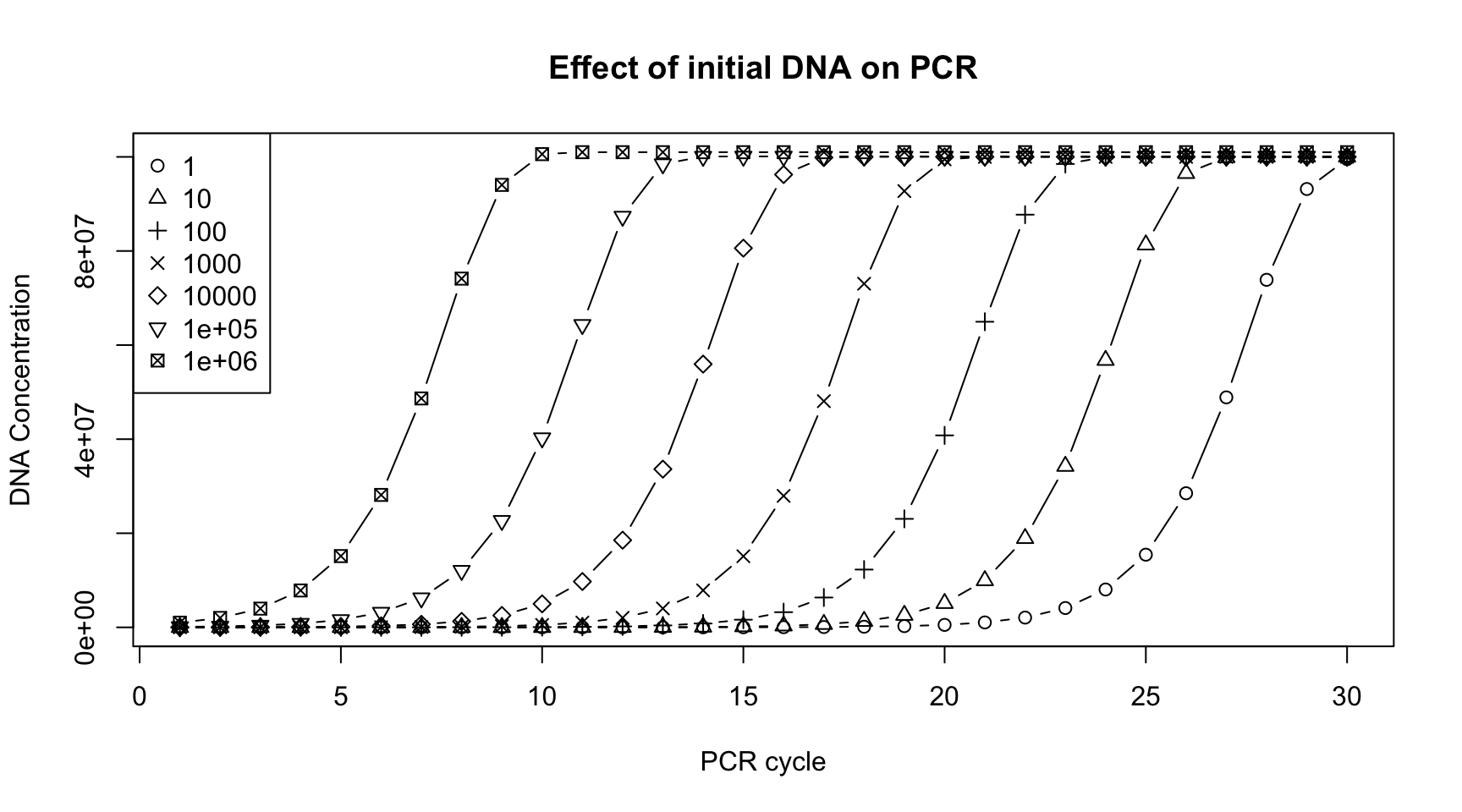
Please write the code (not a function) to make a vector called CT.
The vector CT contains the half value of each column of the data frame conc.
You can use the function locate_half() from Homework 4.
plot(initial_dna ~ CT, log="y")
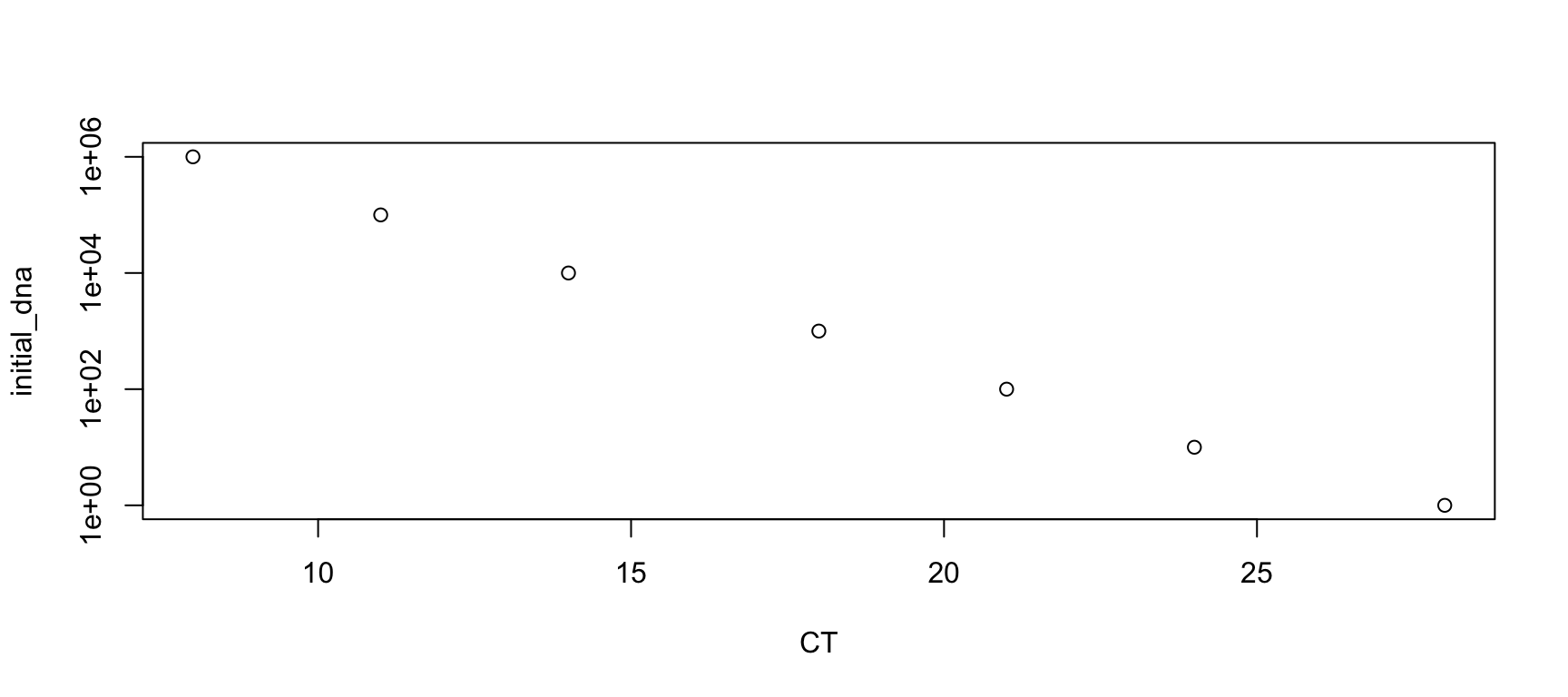 We conclude that the
We conclude that the CT value can be used to predict the initial concentration of DNA.