Searching NCBI has much more options than Google
(do you know Google options?)
By default the query text is searched in any part of any database
But you can specify the fields where you are looking for
- Title of a paper
- author
- date
- taxonomic id
December 10, 2019
Searching NCBI has much more options than Google
(do you know Google options?)
By default the query text is searched in any part of any database
But you can specify the fields where you are looking for
protease NOT hiv1[organism]1000:2000[slen]Mus musculus[organism] AND biomol_mrna[properties]10000:100000[mlwt]src specimen voucher[properties]all[filter] NOT environmental sample[filter] NOT metagenomes[orgn]That is a very good question
Each database has its own fields
Should this be a Homework?
genome assemblies
genes from completely sequenced genomes and that have an active research community to contribute gene-specific data
sequence and map data from the whole genomes. The genomes represent both completely sequenced genomes and those with sequencing in-progress
(Expressed Sequence Tag) sequences from GenBank. Typically short single-pass reads from cDNA libraries generated in survey projects
(Genome Survey Sequence) from GenBank. These are the genomic equivalent of EST records
Apart from sequence data in the EST and GSS divisions, this database contains all the sequence data from GenBank
sequencing data from the next generation sequencing platforms
names and phylogenetic lineages of the more than 350,000 species that have molecular data in the NCBI databases
(Medical Subject Headings) controlled vocabulary and classification system (ontology) used for indexing articles in PubMed. MeSH terminology provides a consistent way to retrieve information that may use different terminology for the same concepts
amino acid sequences created from the translations of coding regions provided on nucleotide records in GenBank, also imported from the outside protein-only data sources (PIR, UniProtKB/Swiss-Prot, Protein Research Foundation, Protein Data Bank)
collection of related protein sequences (clusters) consisting of Reference Sequence proteins that are encoded by complete prokaryotic genomes as well those encoded eukaryotic organelle plasmids and genomes.
protein domains represented by sequence alignments and profiles for protein domains conserved in molecular evolution. It also includes alignments of the domains to known three-dimensional protein structures in the MMDB database.
Molecular Modeling Database (MMDB) contains experimental data from crystallographic and NMR structure determinations. The data for MMDB are obtained from the Protein Data Bank (PDB)
automatically generated sets of homologous genes and their corresponding mRNA, genomic, and protein sequence data from selected eukaryotic organisms.
(Single Nucleotide Polymorphism) database is a central repository for single nucleotide polymorphisms, microsatellites, and small-scale insertions and deletions
complete and incomplete (in-progress) large-scale molecular projects including genome sequencing and assembly, transcriptome, metagenomic, annotation, expression and mapping projects.
contains descriptions of biological source materials used in studies that have data in other NCBI molecular databases such as Assembly, Nucleotide and SRA.
interacting sets of biomolecules involved in metabolic and signaling pathways, disease states, and other biological processes
full-text books that can be searched online and that are linked to PubMed records
database of static NCBI web pages, documentation, and online tools
records for books, journals, audiovisuals, computer software, electronic resources, and other materials in the National Library of Medicine (NLM) collections
(Database of Genotypes and Phenotypes) results of studies on the interaction of genotype and phenotype
(Database of Genomic Structural Variation) contains information about large-scale genomic variation, including large insertions, deletions, translocations and inversions
curated gene expression and molecular abundance data sets from the Gene Expression Omnibus (GEO) repository of microarray data
individual gene expression and molecular abundance profiles assembled from the Gene Expression Omnibus (GEO) repository
nucleic acid reagents designed for use in a wide variety of biomedical research applications including genotyping, gene expression studies, SNP discovery, genome mapping, and gene silencing
database of citations and abstracts for biomedical literature from MEDLINE and additional life science journals
(PMC) is the U.S. National Library of Medicine's digital archive of life sciences journal literature. PMC contains full-text manuscripts deposited by authors or articles provided by the publisher
Entrez queries can be single words, short phrases, sentences, database identifiers, gene symbols, or names
AND: Finds documents that contain terms on both sides of the operator terms, the intersection of both searches.
OR: Finds documents that contain either term, the union of both searches.
NOT: Finds documents that contain the term on the left but not the term on the right of the operator, the subtraction of the right hand search from the one on the left
AND must be in uppercase. It is recommended to also use uppercase for OR and NOT
operators are processed left-to-right
promoters OR response elements NOT human AND mammals
Parenthesis can be used to control the evaluation order
g1p3 AND (response element OR promoter)
" are important[]Each database page includes an Advanced Search option
Certain fields can accept ranges of values
low and high numbers are entered with a colon “:” between them followed by the field
110:500[Sequence Length] 2015/3/1:2016/4/30[Publication Date]
The Clipboard is a temporary place on the NCBI website to save records.
My Collections that is a part of the My NCBI service is a more permanent place to save records.
You need to create an NCBI account to use My NCBI. It is easy and free
There are two major kinds of relationships in the Entrez system:
Combining neighbors and hard links can be an especially effective method for navigating across data and finding the most useful information
When we design molecular biology experiments, or when we analyze their results, we need to use several tools in chain
Today we are going to see an example using the NCBBI website

Our challenge is to find proteins in legumes having F-Box and WD-40 domains
We need an official definition of each domain
Using NCBI Conserved Domain Architecture Retrieval Tool (https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi)
Use the query:
[pfam00646,pfam00400]
to look for proteins that contain both domains in the specified order
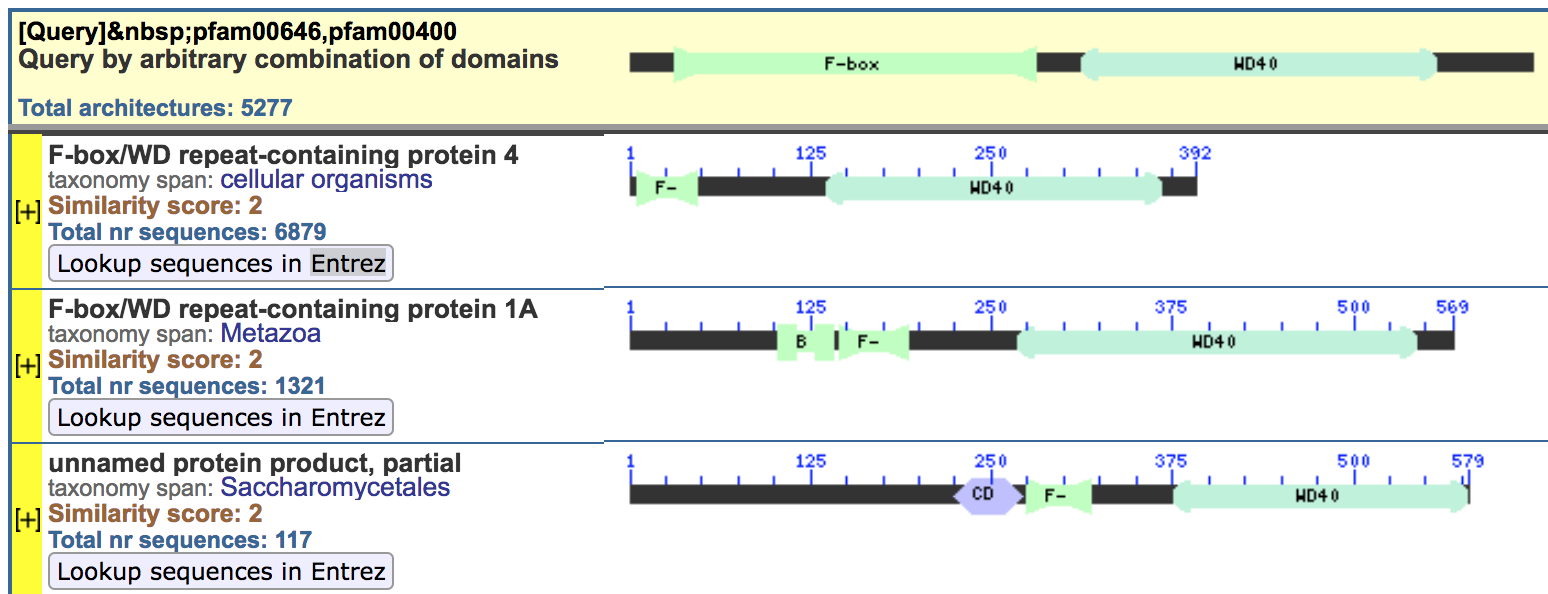
Most of times is a good idea to check the Taxonomy database
Each sequence on GenBank is tagged with a taxon id
Using taxid is more precise than using common names
For example, a protein from human can be labeled “95% similar to mouse”
Is that a human or a mouse protein?
For your convenience you can download the sequences
In this case we only need accession ids
Now we use BLAST to find other proteins in legumes similar to the ones we have
Notice that CDART only has some proteins pre-processed. New sequences take time to be processed
How would you do that?
It is essential that your protocol can be replicated
It is a very good idea to save the search strategy in a file
It is also wise to save the output in a text file
Separate by tab or by comma
How many new proteins we find?
Do all of them have the good domains?
Let’s use CDD again, this time looking for motifs on the new proteins \[protein \to\{domains\}\] (the first time was \(domain\to\{proteins\}\))
And takes a lot of time
It is easy to make mistakes
It is hard to replicate
Can we do it automatically?
ESearch -> ESummary; ESearch -> EFetch; EPost -> ESummary; EPost -> EFetch; ESearch -> ELink; EPost -> ELink; EPost -> ESearch; ELink -> ESearch; ESearch -> ELink -> ESummary; ESearch -> ELink -> EFetch; EPost -> ESearch -> ESummary; EPost -> ESearch -> EFetch; EPost -> ELink -> ESearch -> ESummary; EPost -> ELink -> ESearch -> EFetch;
There are Entrez libraries for most languages
For example in R it is called rentrez
There is a command line version, and versions for all major computer languages
query subject identity positives length 1 ACU21521.1 XP_003538059.1 100.000 100.00 291 2 ACU21521.1 XP_020225874.1 87.542 92.26 297 3 ACU21521.1 XP_007149035.1 87.629 94.16 291 4 ACU21521.1 XP_017425721.1 86.254 92.10 291 5 ACU21521.1 XP_006591036.1 88.660 88.66 291 6 ACU21521.1 XP_014501961.1 85.223 91.07 291 mismatches gaps q.start q.end s.start s.end evalue 1 0 0 1 291 54 344 0 2 31 1 1 291 52 348 0 3 36 0 1 291 50 340 0 4 40 0 1 291 50 340 0 5 0 1 1 291 54 311 0 6 43 0 1 291 50 340 0 score q.gi q.ref s.gi s.ref 1 612 255642515 ACU21521.1 356539142 XP_003538059.1 2 545 255642515 ACU21521.1 1150166268 XP_020225874.1 3 545 255642515 ACU21521.1 593697106 XP_007149035.1 4 526 255642515 ACU21521.1 1044577906 XP_017425721.1 5 526 255642515 ACU21521.1 571488796 XP_006591036.1 6 522 255642515 ACU21521.1 950979929 XP_014501961.1
These are the genbank ids of all the proteins found
[1] "255642515" "1045396645" "1045375294" [4] "1044582125" "1044577908" "1044577906" [7] "1021583843" "1021558720" "1012361995" [10] "1012338638" "1012260727" "1012202223" [13] "965665445" "965609789" "571507141" [16] "571496646" "571488798" "571488796" [19] "356539142" "356501332" "950995503" [22] "950979935" "950979929" "950930754" [25] "947065573" "357493575" "357458443" [28] "920699279" "920691060" "502169256" [31] "502098169" "502090906" "734430373" [34] "734416564" "593701573" "593697106" [37] "593562324" "388522749" "1150166268" [40] "1150166270" "1117517859" "1012225626" [43] "1150166272" "1117517861" "1012225630" [46] "1012225634" "1150128621" "1021534275" [49] "1117375272" "593489431" "1150094071" [52] "1044545110" "1117563883" "571553627" [55] "955389649" "922350178" "571553636" [58] "922329305" "922350180" "1044556548" [61] "1044556546" "1044557823" "1044557821" [64] "1044557819" "951025059" "1044557829" [67] "951025065" "1044557825" "1044557827" [70] "951025063" "571482571" "571482569" [73] "593689820" "571482575" "571482573" [76] "1150095614" "502183121" "1150095616" [79] "828339994" "502183133" "502183129" [82] "922400539" "357439909" "828339999" [85] "502183147" "502183143" "502183138" [88] "571440442" "571440440" "571440444" [91] "356500353" "1117375772" "1117375759" [94] "1117375765" "1117375785" "1117375778" [97] "1117546227" "1117546205" "1117375768" [100] "1117375787" "1117375775" "1117375781" [103] "1117375790" "955307577" "955307575" [106] "1117546230" "1117546224" "955307580" [109] "951017511" "593689824" "1117342058" [112] "1117342051" "1117342038" "356537561" [115] "1117342048" "1117342055" "1021550847" [118] "1021550849" "1150095590" "1012214348" [121] "1044553727" "502103542" "828298200" [124] "1150095620" "951017515" "571440447" [127] "593689826" "955384590" "571546627" [130] "950962514" "950962510" "356567862" [133] "571546623" "1044534197" "1044534201" [136] "357505281" "593441102" "357505277" [139] "593689822" "1150093585" "1021530819" [142] "571474527" "1012196136" "1021530815" [145] "1012196130" "1021530817" "1012196133" [148] "502132291" "502132289" "571509404" [151] "356552535" "1150118542" "1150118544" [154] "502132293" "1044523131" "828313695" [157] "1150118546" "356552537" "1150118548" [160] "356503387" "571474529" "1012184691" [163] "1012184695" "357509397" "1150136062" [166] "955314277" "593562195" "1044582959" [169] "950983855" "356553464" "356499483" [172] "1150127909" "828296784" "357495283" [175] "1117372388" "1012199286" "1021533997"
domains <- entrez_link(dbfrom = "protein", id=proteins,
by_id=TRUE, db="cdd")
doms <- lapply(domains,
function(m) m$links$protein_cdd_concise_2)
udomains <- unique(unlist(doms))
udomains
[1] "356469" "338561" "197608" "334189" "238121" [6] "225201" "351360" "337084" "294672" "312941" [11] "197891"
dom.summary <- entrez_summary("cdd", udomains)
dom.title <- extract_from_esummary(dom.summary, "title")
prot.domains <- sapply(doms,
function(d) paste(dom.title[d], collapse=" "))
unique(prot.domains)
[1] "WD40 F-box-like" [2] "WD40 FBOX" [3] "F-box-like WD40" [4] "WD40 F-box" [5] "WD40 F-box-like WD40" [6] "" [7] "WD40" [8] "WD40 WD40 F-box-like" [9] "SGNH_hydrolase WD40 F-box-like" [10] "WD40 LisH Dyp_perox" [11] "WD40 LisH" [12] "WD40 Med15 LisH" [13] "WD40 LisH Med15" [14] "F-box" [15] "WD40 Amelogenin LisH" [16] "WD40 WD40"
wd40.id <- names(dom.title)[dom.title=="WD40"]
fbox.id <- names(dom.title)[dom.title %in%
c("F-box", "F-box-like")]
has.wd40 <- sapply(doms,
function(d) length(intersect(d, wd40.id))>0)
has.fbox <- sapply(doms,
function(d) length(intersect(d, fbox.id))>0)
has.both <- has.fbox & has.wd40
table(has.both)
has.both FALSE TRUE 134 43
prot.seq <- entrez_fetch(db="protein", rettype = "fasta",
id=proteins[has.both])
write(prot.seq, file="ncbi-proteins-wd40-fbox.faa")
messn <- entrez_link(dbfrom = "protein", id=proteins,
by_id=TRUE, linkname="protein_nuccore_mrna")
messn <- sapply(messn,
function(m) m$links$protein_nuccore_mrna)
messn.seq <- entrez_fetch(db="nuccore", rettype = "fasta",
id=messn[has.both])
write(messn.seq, file="ncbi-messngr-wd40-fbox.fna")
genes <- entrez_link(dbfrom = "protein", by_id=TRUE,
id=proteins[has.both], linkname="protein_gene")
ugenes <- sapply(genes, function(m) m$links$protein_gene)
geoprof <- entrez_link(dbfrom = "gene", id=ugenes,
by_id=TRUE, linkname="gene_geoprofiles")
profiles <- lapply(geoprof,
function(m) m$links$gene_geoprofiles)
ugeoprof <- unique(unlist(profiles))
geoprofiles_gds <- entrez_link(dbfrom = "geoprofiles",
id=ugeoprof, linkname="geoprofiles_gds")
gene_pubmed <- entrez_link(dbfrom = "gene", id=ugenes,
by_id=TRUE, linkname="gene_pubmed")
upubmed <- unique(unlist(lapply(gene_pubmed,
function(m) m$links$gene_pubmed)))
recs <- entrez_fetch(db="pubmed", id=upubmed,
rettype="xml")
papers <- parse_pubmed_xml(recs)
sonuc <- data.frame(proteins,
name=pnames[proteins],
messn=sapply(messn,
function(m) ifelse(is.null(m),"",m[1])),
domains=prot.domains,
has.fbox, has.wd40, has.both,
stringsAsFactors = FALSE)