March 7, 2018
Judo
Judo and Aikido

Judo and Aikido

- In both sports you loose when you fall
- Each training session starts with training how to fall
- You train to win and to fail
- Because failure is part of the game
- You will not always win
- Learn to fail without hurting getting injured
- Turn your failure in the beginning of the next match
Failing is part of learning
“Perfect is the Enemy of Good”

Do something bad, then improve it
Then improve it again
Like Michelangelo sculptures
Number of rabbits
We represent the number of pairs of rabbits on month n with the function fib(n).
This function has the following rules:
fib(0) = 0fib(1) = 1fib(n) = fib(n-1) + fib(n-2)
Your task is to write a recursive function in R that has input n and output fib(n).
fib() is a recursive function
Recursive means that the function calls itself
It is like the factorial example
facto <- function(n) {
if(n <= 1) {
ans <- 1
} else {
m <- n-1
ans <- n * facto(m)
}
return(ans)
}
Let’s adapt it to Fibonacci
- The name changes from
factointofib - There are two exit conditions
fib(0) = 0meansif(n==0) {ans <- 0}fib(1) = 1meansif(n==1) {ans <- 1}
A simple (incomplete) solution
# author: I CAN NOT DO THİS
fib <- function(n) {
if(n==0) {
ans <- 0
if(n==1) {
ans <- 1
} else {
ans <- fib(n-1) + fib(n-2)
}
return(ans)
}
First, use indentation to see structure
Indentation: use spaces to see the structure
fib <- function(n) {
if(n==0) {
ans <- 0
if(n==1) {
ans <- 1
} else {
ans <- fib(n-1) + fib(n-2)
}
return(ans)
}
Now you can see that you are missing a { before the second if, and a } at the end
Close braces in the correct place
fib <- function(n) {
if(n==0) {
ans <- 0
}
if(n==1) {
ans <- 1
} else {
ans <- fib(n-1) + fib(n-2)
}
return(ans)
}
Use the debugger to see what happens when n is 0
(use h key)
Did you found the missing part?
fib <- function(n) {
if(n <= 0){
ans <- 0
} else if(n <= 1) {
ans <- 1
} else {
ans <- fib(n-1) + fib(n-2)
}
return(ans)
}
(use h key)
Other details
- This:
n<=0is a comparison. Result is logic This:
n<-0is an assignment. Result is numeric- This:
n==0is a comparison. Result is logic This:
n=0is an assignment. Result is numeric
Make it Simple
The cases n==0 and n==1 can be put together
fib <- function(n) {
if(n <= 1){
ans <- n
} else {
ans <- fib(n-1) + fib(n-2)
}
return(ans)
}
This is optional, but nice
(use h key)
Stick-person
Draw a head
turtle_right(90) turtle_forward(4.5) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(4.5) turtle_right(90)

Arms
# upper body turtle_forward(9) turtle_right(180) turtle_forward(3) # arms turtle_left(90) turtle_forward(9) turtle_right(90) turtle_right(90) turtle_forward(18) turtle_left(180) turtle_forward(9) turtle_left(90)

Draw the first leg
turtle_forward(10) turtle_forward(5) # first leg turtle_right(40) turtle_forward(10) turtle_forward(3) turtle_left(90) turtle_forward(3) turtle_left(180) turtle_forward(3) turtle_right(40) turtle_right(50) turtle_forward(13)

Draw the second leg
# get back to initial angle turtle_right(40) turtle_right(90) turtle_left(10) turtle_right(20) # second leg turtle_left(5) turtle_left(40) turtle_forward(13) turtle_left(90) turtle_forward(3) turtle_hide()

Can we simplify it?
Replace repeated pattern by loop
turtle_right(90) turtle_forward(4.5) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(9) turtle_left(90) turtle_forward(4.5) turtle_right(90)
turtle_right(90)
turtle_forward(4.5)
for(i in 1:3) {
turtle_left(90)
turtle_forward(9)
}
turtle_left(90)
turtle_forward(4.5)
turtle_right(90)
(use h key)
The size can be variable
turtle_right(90)
turtle_forward(4.5)
for(i in 1:3) {
turtle_left(90)
turtle_forward(9)
}
turtle_left(90)
turtle_forward(4.5)
Let’s say size=9 (for now)
turtle_right(90)
turtle_forward(size/2)
for(i in 1:3) {
turtle_left(90)
turtle_forward(size)
}
turtle_left(90)
turtle_forward(size/2)
(use h key)
Simplify and generalize the arms
# upper body turtle_forward(9) turtle_right(180) turtle_forward(3) # arms turtle_left(90) turtle_forward(9) turtle_right(90) turtle_right(90) turtle_forward(18) turtle_left(180) turtle_forward(9) turtle_left(90)
# upper body turtle_forward(size*2/3) # arms turtle_right(90) turtle_forward(size) turtle_right(180) turtle_forward(size*2) turtle_left(180) turtle_forward(size) turtle_left(90)
First leg
turtle_forward(10) turtle_forward(5) # first leg turtle_right(40) turtle_forward(10) turtle_forward(3) turtle_left(90) turtle_forward(3) turtle_left(180) turtle_forward(3) turtle_right(40) turtle_right(50) turtle_forward(13)
turtle_forward(15) # first leg turtle_right(40) turtle_forward(13) turtle_left(90) turtle_forward(3) turtle_left(180) turtle_forward(3) turtle_right(90) turtle_forward(13)
Use variable size
turtle_forward(15) turtle_right(40) # first leg turtle_forward(13) turtle_left(90) turtle_forward(3) turtle_left(180) turtle_forward(3) turtle_right(90) turtle_forward(13)
turtle_forward(size*15/9) turtle_right(40) # first leg turtle_forward(size*13/9) turtle_left(90) turtle_forward(size/3) turtle_left(180) turtle_forward(size/3) turtle_right(90) turtle_forward(size*13/9)
Clear code on second leg
# get back to initial angle turtle_right(40) turtle_right(90) turtle_left(10) turtle_right(20) # first leg turtle_left(5) turtle_left(40) turtle_forward(13) turtle_left(90) turtle_forward(3)
\(40+90-10+20 = 140\)
# get back to initial angle turtle_right(140) # first leg turtle_left(45) turtle_forward(13) turtle_left(90) turtle_forward(3)
Abstraction & decomposition
draw_person <- function(size) {
draw_head(size*1.2)
turtle_left(180)
turtle_forward(size)
turtle_left(90)
draw_arm(size*1.5)
turtle_left(180)
draw_arm(size*1.5)
turtle_left(90)
turtle_forward(size*2)
turtle_left(20)
draw_leg(size*2)
turtle_right(40)
draw_leg(size*2)
}
This is the main function.
It is my part.
I can change it.
You should not change it.
Instead, you have to provide functions for head, arms and legs.
Body parts
draw_head <- function(size) {
}
draw_arm <- function(size) {
}
draw_leg <- function(size) {
}
This is a Contract
Each part commits to do something
Each part makes a promise
We promise to leave the turtle in the same position as we received it
Let’s start with the arms
You should always start with the easy parts
The easy part may not be the first part
We receive the turtle pointing in the arm direction
We must leave the turtle in the same place and same angle
draw_arm <- function(size) {
turtle_forward(size)
turtle_backward(size)
}
From arms to legs
draw_arm <- function(size) {
turtle_forward(size)
turtle_backward(size)
}
draw_leg <- function(size) {
turtle_forward(size)
turtle_left(90)
turtle_forward(size/3)
turtle_backward(size/3)
turtle_right(90)
turtle_backward(size)
}
Undoing
Notice that to undo something you have to undo each part in reverse order
You put your socks first, then your shoes
To undo you first “un-put” your shoes, then your socks
In general
\[Undo(A,B,C) = Undo(C), Undo(B), Undo(A)\]
There is another way
Each function has a separate environment with its own variables
We can store position and angle at the start, and reset them at the end
draw_leg <- function(size) {
old_pos <- turtle_getpos()
old_angle <- turtle_getangle()
turtle_forward(size)
turtle_left(90)
turtle_forward(size/3)
turtle_setangle(old_angle)
turtle_setpos(old_pos[1], old_pos[2])
}
This is useful when the drawing is complex. Keep it in mind
Separation of concerns

In this case we separated the big problem on independent parts. That allows us to change each part without affecting the others, as long as we keep our promises.
For example, you can change the position of the hands, the shape of the head and hands, an others. The code is in stick-person-2.R.
Figures from DNA
Candidatus Carsonella ruddii has the smallest genome known
Candidatus Carsonella ruddii is a obligate symbiont of Pachpsylla venusta.
There is much controversy over whether this is a living cell or simply an organelle as it is missing genes needed for living independently.
Published as “The 160-kilobase genome of the bacterial endosymbiont Carsonella.” https://www.ncbi.nlm.nih.gov/pubmed/17038615
C. ruddii DNA looks like this
and many more lines
ATGAATACTATATTTTCAAGAATAACACCATTAGGAAATGGTACGTTATGTGTTATAAGAATTTCTGGAA AAAATGTAAAATTTTTAATACAAAAAATTGTAAAAAAAAATATAAAAGAAAAAATAGCTACTTTTTCTAA ATTATTTTTAGATAAAGAATGTGTAGATTATGCAATGATTATTTTTTTTAAAAAACCAAATACGTTCACT GGAGAAGATATAATCGAATTTCATATTCACAATAATGAAACTATTGTAAAAAAAATAATTAATTATTTAT TATTAAATAAAGCAAGATTTGCAAAAGCTGGCGAATTTTTAGAAAGACGATATTTAAATGGAAAAATTTC TTTAATAGAATGCGAATTAATAAATAATAAAATTTTATATGATAATGAAAATATGTTTCAATTAACAAAA AATTCTGAAAAAAAAATATTTTTATGTATAATTAAAAATTTAAAATTTAAAATAAATTCTTTAATAATTT GTATTGAAATCGCAAATTTTAATTTTAGTTTTTTTTTTTTTAATGATTTTTTATTTATAAAATATACATT TAAAAAACTATTAAAACTTTTAAAAATATTAATTGATAAAATAACTGTTATAAATTATTTAAAAAAGAAT TTCACAATAATGATATTAGGTAGAAGAAATGTAGGAAAGTCTACTTTATTTAATAAAATATGTGCACAAT ATGACTCGATTGTAACTAATATTCCTGGTACTACAAAAAATATTATATCAAAAAAAATAAAAATTTTATC TAAAAAAATAAAAATGATGGATACAGCAGGATTAAAAATTAGAACTAAAAATTTAATTGAAAAAATTGGA ATTATTAAAAATATAAATAAAATTTATCAAGGAAATTTAATTTTGTATATGATTGATAAATTTAATATTA AAAATATATTTTTTAACATTCCAATAGATTTTATTGATAAAATTAAATTAAATGAATTAATAATTTTAGT TAACAAATCAGATATTTTAGGAAAAGAAGAAGGAGTTTTTAAAATAAAAAATATATTAATAATTTTAATT TCTTCTAAAAATGGAACTTTTATAAAAAATTTAAAATGTTTTATTAATAAAATCGTTGATAATAAAGATT TTTCTAAAAATAATTATTCTGATGTTAAAATTCTATTTAATAAATTTTCTTTTTTTTATAAAGAATTTTC ATGTAACTATGATTTAGTGTTATCAAAATTAATTGATTTTCAAAAAAATATATTTAAATTAACAGGAAAT TTTACTAATAAAAAAATAATAAATTCTTGTTTTAGAAATTTTTGTATTGGTAAATGAATATTTTTAATAT AATTATTATTGGAGCAGGACATTCTGGTATAGAAGCAGCTATATCTGCATCTAAAATATGTAATAAAATA
We can use the Turtle to see the DNA
- For each nucleotide
- if the nucleotide is
A, turtle moves left - if the nucleotide is
T, turtle moves right - if the nucleotide is
G, turtle moves up - if the nucleotide is
C, turtle moves down
- if the nucleotide is
The Turtle walks the DNA sequence
library(TurtleGraphics)
turtle_init(mode = "clip")
turtle_hide()
for(i in 1:1000) {
if(dna[i]=="A") {
turtle_setangle(90)
} else if(dna[i]=="T") {
turtle_setangle(270)
} else if(dna[i]=="G") {
turtle_setangle(0)
} else if(dna[i]=="C") {
turtle_setangle(180)
}
turtle_forward(1)
}
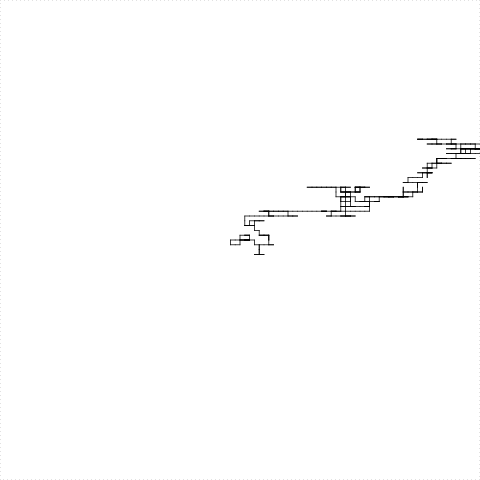
But the turtle is too slow
it is a turtle, after all
- We can do the graphic without the turtle
- We can use a normal plot, like the last semester
- We can draw line segments with
segments(x0, y0, x1, y1) - We start with an empty plot, without labels
- Option
type="n" - Option
ann=FALSE xandygive horizontal and vertical range
- Option
- We reduce margin using
par(mar=...)
Read the help of segments, par and plot.default
Same without Turtle
par(mar=c(2, 2, 0.3, 0.3))
plot(x=c(-500,2500), y=c(-700,1100),
type = "n", ann=FALSE)
x <- y <- 0
for(i in 1:length(dna)) {
if(dna[i]=="A") {
segments(x, y, x+1, y)
x <- x+1
} else if(dna[i]=="T") {
segments(x, y, x-1, y)
x <- x-1
} else if(dna[i]=="G") {
segments(x, y, x, y+1)
y <- y+1
} else if(dna[i]=="C") {
segments(x, y, x, y-1)
y <- y-1
}
}

Scientific questions about DNA-walk
- Why do we got this shape?
- What is the shape for other organisms?
- Do you think this shape is random?
- Do you think there is some logic? some structure?
- What are the minimum and maximum of each axis?
How can you answer these questions?
Molecular Biology and Sequences
DNA
- A big molecule, but not too complex. It is a polymer
- All made of only 4 pieces (pattern again!)
Proteins
Looks more complex, but it is still a polymer
DNA and proteins are easy to model
Despite being large molecules, DNA and proteins are made with pieces of only a few types
DNA is made of four bases. We can represent it with four letters
AGCTTTTCATTCTGACTGCAACGGGCAATATGTCTCTGTGTGGATTAAAAAAAGAGTGTCTGATAGCAGC
Proteins have 20 aminoacids and three stop codons. We can represent them with 23 letters.
MKRISTTITTTITITTGNGAG
Sequence data
In molecular biology we often work with sequences
- DNA sequences use 4 letters to represent the nucleotides in one of the two strands
- Protein sequences use 20 letters to represent the amino-acids, from amino to carboxyl terminal
- Other sequences are sometime used:
- RNA,
- DNA with ambiguous nucleotides,
- amino-acid sequences with stop codons
Sequence data is digital data
The main reason why computing is useful for molecular biology
- DNA is discrete data
- Either “A”, “C”, “G” or “T”
- No middle-values
- All other things we measure are in a range
- Like temperature, concentration, expression
- We can forget all the chemistry and work with symbols
Sequences are stored in FASTA format
There are several ways to store DNA or protein data
Most of the times they are stored in FASTA format
FASTA files are text files, with some rules
You should never use word to store sequences
Example FASTA file
AP009180.1 Candidatus Carsonella ruddii PV DNA, complete genome ATGAATACTATATTTTCAAGAATAACACCATTAGGAAATGGTACGTTATGTGTTATAAGAATTTCTGGAA AAAATGTAAAATTTTTAATACAAAAAATTGTAAAAAAAAATATAAAAGAAAAAATAGCTACTTTTTCTAA ATTATTTTTAGATAAAGAATGTGTAGATTATGCAATGATTATTTTTTTTAAAAAACCAAATACGTTCACT GGAGAAGATATAATCGAATTTCATATTCACAATAATGAAACTATTGTAAAAAAAATAATTAATTATTTAT TATTAAATAAAGCAAGATTTGCAAAAGCTGGCGAATTTTTAGAAAGACGATATTTAAATGGAAAAATTTC TTTAATAGAATGCGAATTAATAAATAATAAAATTTTATATGATAATGAAAATATGTTTCAATTAACAAAA AATTCTGAAAAAAAAATATTTTTATGTATAATTAAAAATTTAAAATTTAAAATAAATTCTTTAATAATTT GTATTGAAATCGCAAATTTTAATTTTAGTTTTTTTTTTTTTAATGATTTTTTATTTATAAAATATACATT TAAAAAACTATTAAAACTTTTAAAAATATTAATTGATAAAATAACTGTTATAAATTATTTAAAAAAGAAT TTCACAATAATGATATTAGGTAGAAGAAATGTAGGAAAGTCTACTTTATTTAATAAAATATGTGCACAAT ATGACTCGATTGTAACTAATATTCCTGGTACTACAAAAAATATTATATCAAAAAAAATAAAAATTTTATC TAAAAAAATAAAAATGATGGATACAGCAGGATTAAAAATTAGAACTAAAAATTTAATTGAAAAAATTGGA ATTATTAAAAATATAAATAAAATTTATCAAGGAAATTTAATTTTGTATATGATTGATAAATTTAATATTA AAAATATATTTTTTAACATTCCAATAGATTTTATTGATAAAATTAAATTAAATGAATTAATAATTTTAGT TAACAAATCAGATATTTTAGGAAAAGAAGAAGGAGTTTTTAAAATAAAAAATATATTAATAATTTTAATT TCTTCTAAAAATGGAACTTTTATAAAAAATTTAAAATGTTTTATTAATAAAATCGTTGATAATAAAGATT TTTCTAAAAATAATTATTCTGATGTTAAAATTCTATTTAATAAATTTTCTTTTTTTTATAAAGAATTTTC ATGTAACTATGATTTAGTGTTATCAAAATTAATTGATTTTCAAAAAAATATATTTAAATTAACAGGAAAT TTTACTAATAAAAAAATAATAAATTCTTGTTTTAGAAATTTTTGTATTGGTAAATGAATATTTTTAATAT AATTATTATTGGAGCAGGACATTCTGGTATAGAAGCAGCTATATCTGCATCTAAAATATGTAATAAAATA
… and more
FASTA file (amino acids)
>NC_000913.3_prot_NP_414542.1_1 [gene=thrL] [protein=thr operon leader peptide] [protein_id=NP_414542.1] [location=190..255] MKRISTTITTTITITTGNGAG >NC_000913.3_prot_NP_414543.1_2 [gene=thrA] [protein=Bifunctional aspartokinase/homoserine dehydrogenase 1] [protein_id=NP_414543.1] [location=337..2799] MRVLKFGGTSVANAERFLRVADILESNARQGQVATVLSAPAKITNHLVAMIEKTISGQDALPNISDAERI FAELLTGLAAAQPGFPLAQLKTFVDQEFAQIKHVLHGISLLGQCPDSINAALICRGEKMSIAIMAGVLEA RGHNVTVIDPVEKLLAVGHYLESTVDIAESTRRIAASRIPADHMVLMAGFTAGNEKGELVVLGRNGSDYS AAVLAACLRADCCEIWTDVDGVYTCDPRQVPDARLLKSMSYQEAMELSYFGAKVLHPRTITPIAQFQIPC LIKNTGNPQAPGTLIGASRDEDELPVKGISNLNNMAMFSVSGPGMKGMVGMAARVFAAMSRARISVVLIT QSSSEYSISFCVPQSDCVRAERAMQEEFYLELKEGLLEPLAVTERLAIISVVGDGMRTLRGISAKFFAAL ARANINIVAIAQGSSERSISVVVNNDDATTGVRVTHQMLFNTDQVIEVFVIGVGGVGGALLEQLKRQQSW LKNKHIDLRVCGVANSKALLTNVHGLNLENWQEELAQAKEPFNLGRLIRLVKEYHLLNPVIVDCTSSQAV ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENLQNLLNAGDELM KFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDVARKLLILARETGRELELADIE IEPVLPAEFNAEGDVAAFMANLSQLDDLFAARVAKARDEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFK VKNGENALAFYSHYYQPLPLVLRGYGAGNDVTAAGVFADLLRTLSWKLGV >NC_000913.3_prot_NP_414544.1_3 [gene=thrB] [protein=homoserine kinase] [protein_id=NP_414544.1] [location=2801..3733] MVKVYAPASSANMSVGFDVLGAAVTPVDGALLGDVVTVEAAETFSLNNLGRFADKLPSEPRENIVYQCWE RFCQELGKQIPVAMTLEKNMPIGSGLGSSACSVVAALMAMNEHCGKPLNDTRLLALMGELEGRISGSIHY DNVAPCFLGGMQLMIEENDIISQQVPGFDEWLWVLAYPGIKVSTAEARAILPAQYRRQDCIAHGRHLAGF IHACYSRQPELAAKLMKDVIAEPYRERLLPGFRQARQAVAEIGAVASGISGSGPTLFALCDKPETAQRVA DWLGKNYLQNQEGFVHICRLDTAGARVLEN >NC_000913.3_prot_NP_414545.1_4 [gene=thrC] [protein=L-threonine synthase] [protein_id=NP_414545.1] [location=3734..5020] MKLYNLKDHNEQVSFAQAVTQGLGKNQGLFFPHDLPEFSLTEIDEMLKLDFVTRSAKILSAFIGDEIPQE ILEERVRAAFAFPAPVANVESDVGCLELFHGPTLAFKDFGGRFMAQMLTHIAGDKPVTILTATSGDTGAA VAHAFYGLPNVKVVILYPRGKISPLQEKLFCTLGGNIETVAIDGDFDACQALVKQAFDDEELKVALGLNS ANSINISRLLAQICYYFEAVAQLPQETRNQLVVSVPSGNFGDLTAGLLAKSLGLPVKRFIAATNVNDTVP RFLHDGQWSPKATQATLSNAMDVSQPNNWPRVEELFRRKIWQLKELGYAAVDDETTQQTMRELKELGYTS EPHAAVAYRALRDQLNPGEYGLFLGTAHPAKFKESVEAILGETLDLPKELAERADLPLLSHNLPADFAAL RKLMMNHQ >NC_000913.3_prot_NP_414546.1_5 [gene=yaaX] [protein=DUF2502 family putative periplasmic protein] [protein_id=NP_414546.1] [location=5234..5530] MKKMQSIVLALSLVLVAPMAAQAAEITLVPSVKLQIGDRDNRGYYWDGGHWRDHGWWKQHYEWRGNRWHL HGPPPPPRHHKKAPHDHHGGHGPGKHHR >NC_000913.3_prot_NP_414547.1_6 [gene=yaaA] [protein=peroxide resistance protein, lowers intracellular iron] [protein_id=NP_414547.1] [location=complement(5683..6459)] MLILISPAKTLDYQSPLTTTRYTLPELLDNSQQLIHEARKLTPPQISTLMRISDKLAGINAARFHDWQPD FTPANARQAILAFKGDVYTGLQAETFSEDDFDFAQQHLRMLSGLYGVLRPLDLMQPYRLEMGIRLENARG
Reading FASTA in R
To handle sequence data in R, we use the seqinr library
You have to install it once.
install.packages("seqinr")
Then you have to load it on every session
library(seqinr)
