December 20, 2018
Other things to do in Rstudio
Other computer languages
You can use other computer languages besides R, such as python
Just write the language at the beginning ```{python}
This calculates the \(e\) number in Python
x=10**50 e=x for i in range(30): x //= (i+1) e += x print(e)
271828182845904523536028747135266237222570213098336
Other languages
You can draw networks from their text description
digraph { size="4,4"; rankdir=LR; bgcolor="transparent";
A->B; A->C; B->C; B->D; D->E; C->E;}
Visualization of matrices
dim(volcano)
[1] 87 61
volcano[1:10,1:10]
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
[1,] 100 100 101 101 101 101 101 100 100
[2,] 101 101 102 102 102 102 102 101 101
[3,] 102 102 103 103 103 103 103 102 102
[4,] 103 103 104 104 104 104 104 103 103
[5,] 104 104 105 105 105 105 105 104 104
[6,] 105 105 105 106 106 106 106 105 105
[7,] 105 106 106 107 107 107 107 106 106
[8,] 106 107 107 108 108 108 108 107 107
[9,] 107 108 108 109 109 109 109 108 108
[10,] 108 109 109 110 110 110 110 109 109
[,10]
[1,] 100
[2,] 101
[3,] 102
[4,] 103
[5,] 103
[6,] 104
[7,] 105
[8,] 106
[9,] 107
[10,] 108
Image: each values one pixel
Here lower values are red, higher values are white. Like heating metal
image(volcano)
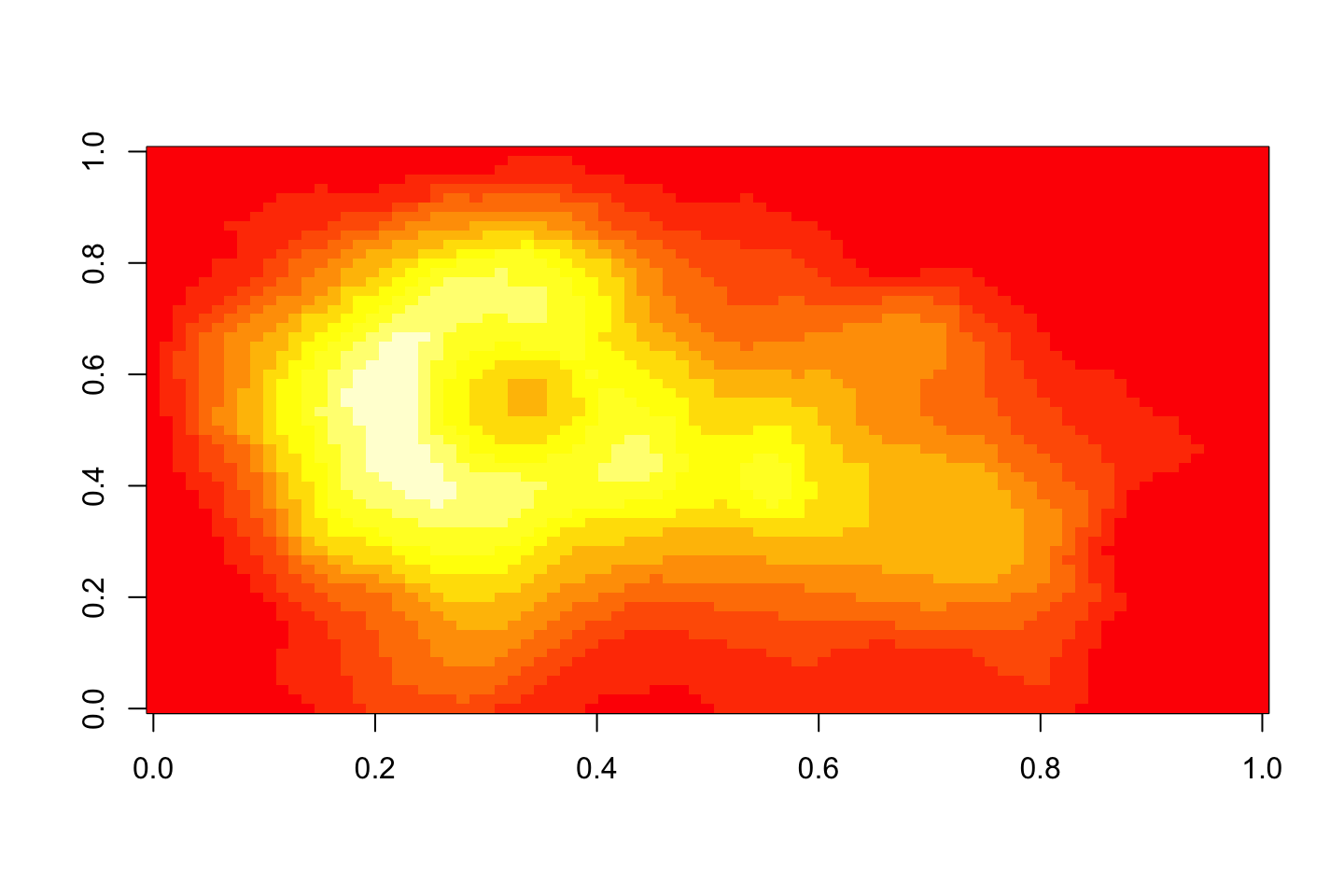
More colors in the same palette
image(volcano, col=heat.colors(100))
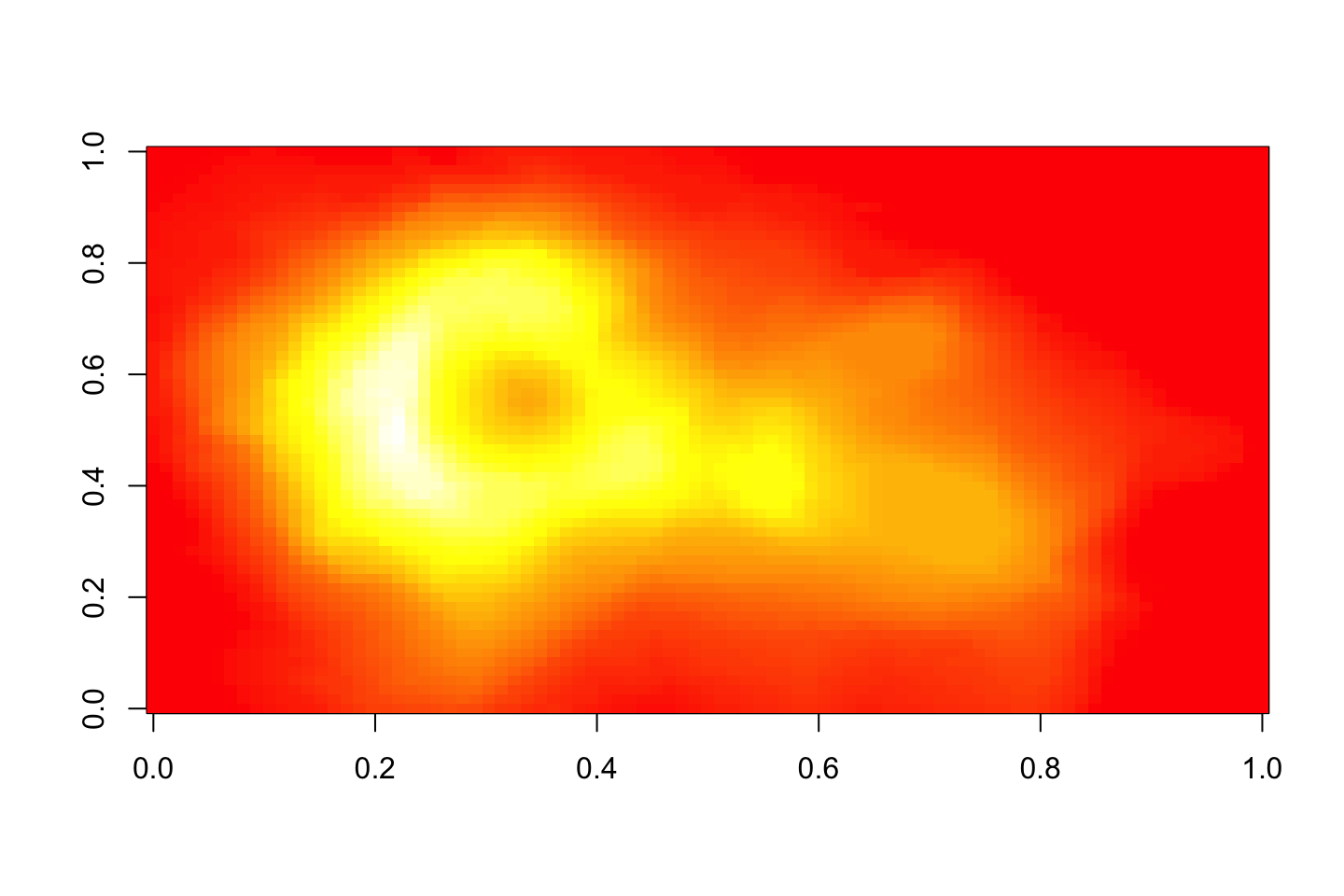
Change color palette
image(volcano, col=gray.colors(100))

Image
image(volcano, col=terrain.colors(100))
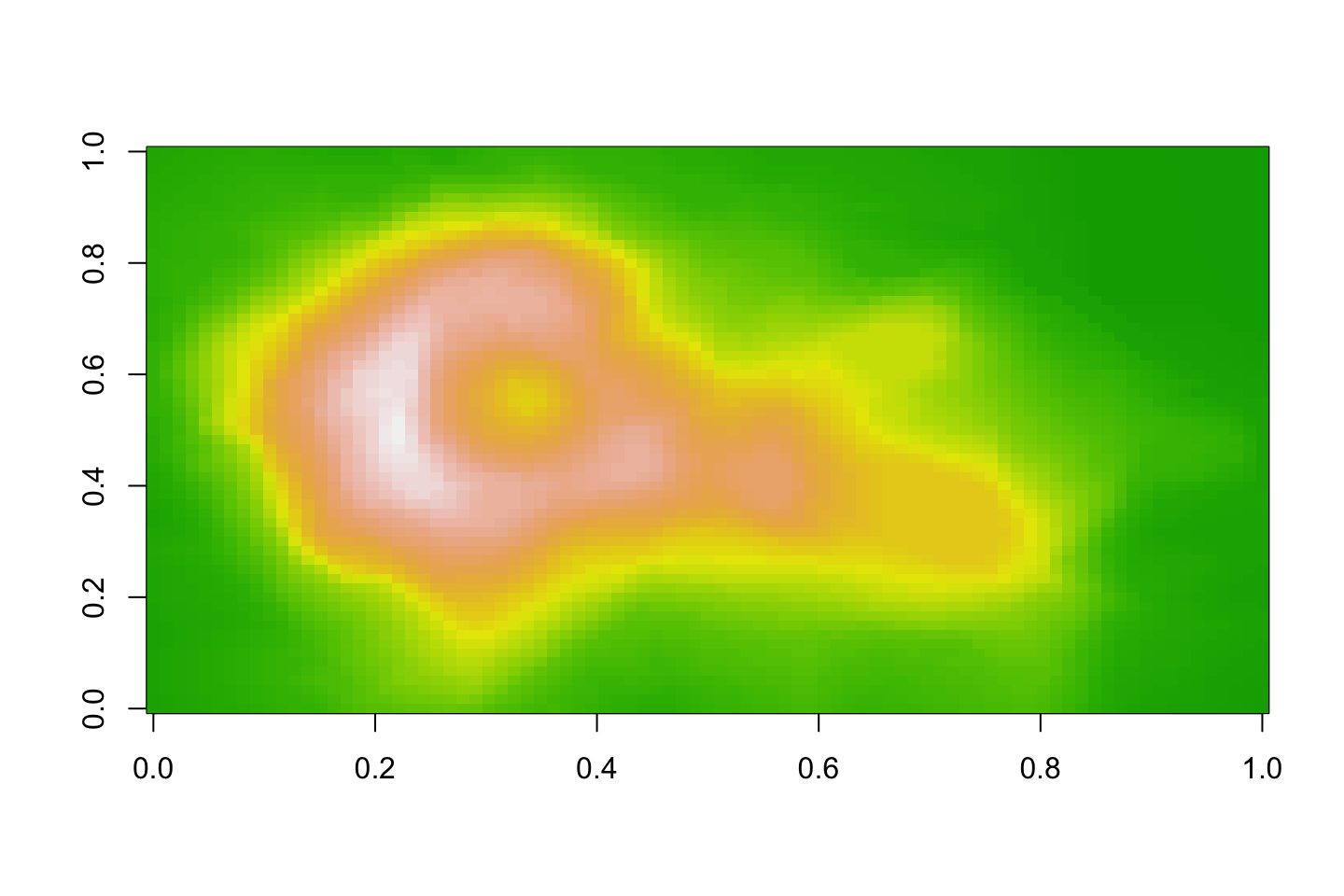
Image
image(volcano, col=topo.colors(100))
Image
image(volcano, col=cm.colors(100))

Contour: like a map
contour(volcano)
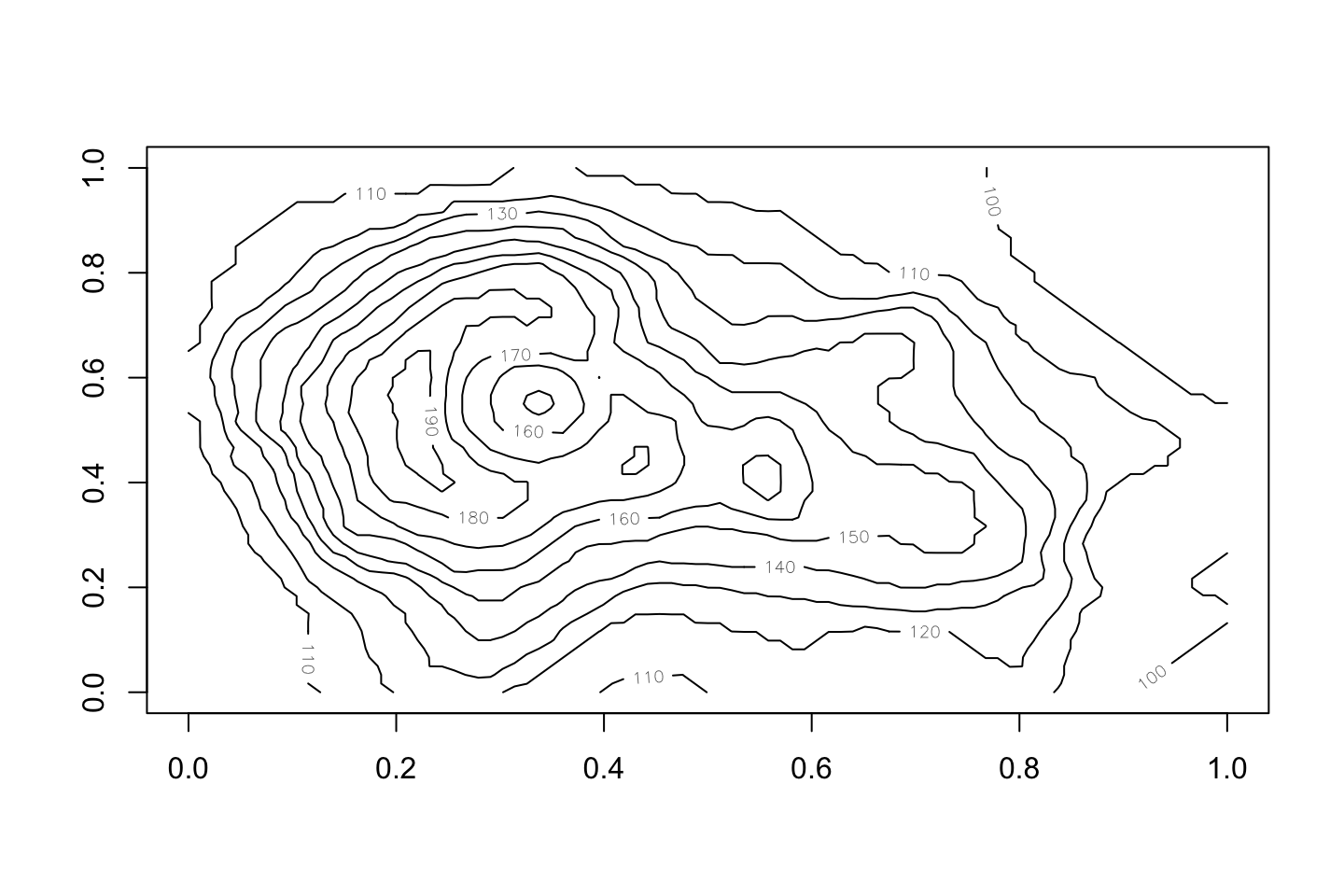
Image and contour
image(volcano, col=terrain.colors(100)) contour(volcano, add=TRUE)
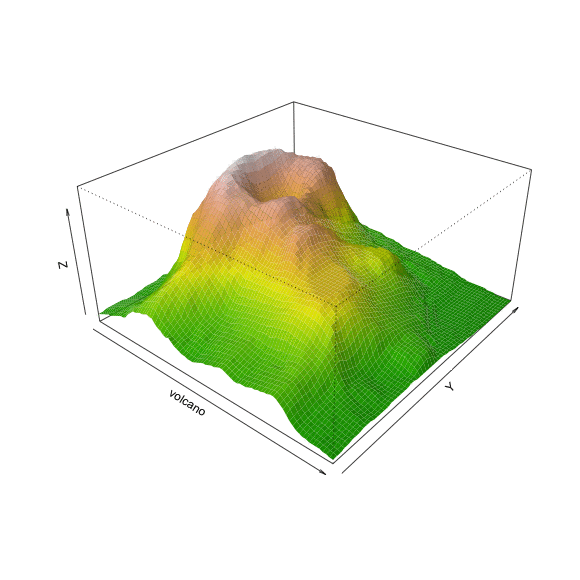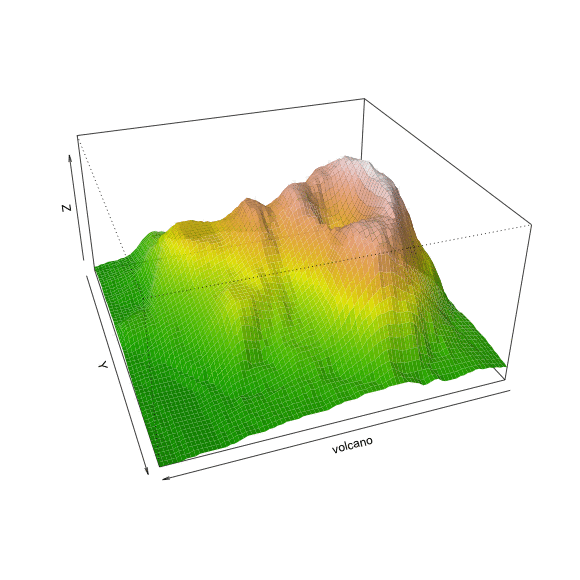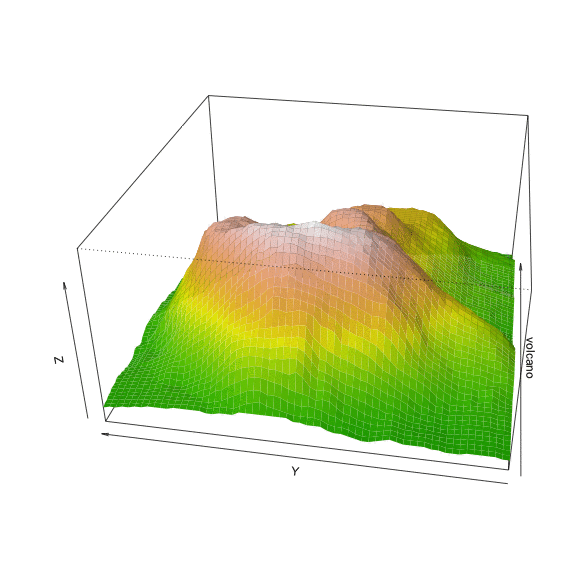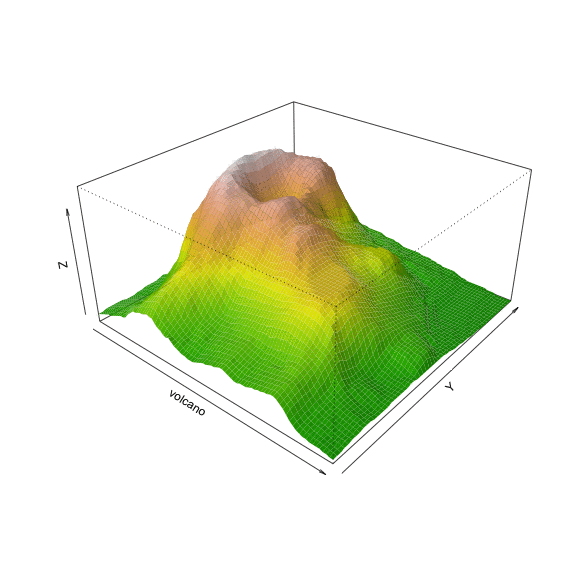
Perspective
persp(volcano)
Perspective
persp(volcano, theta=40, phi=30)
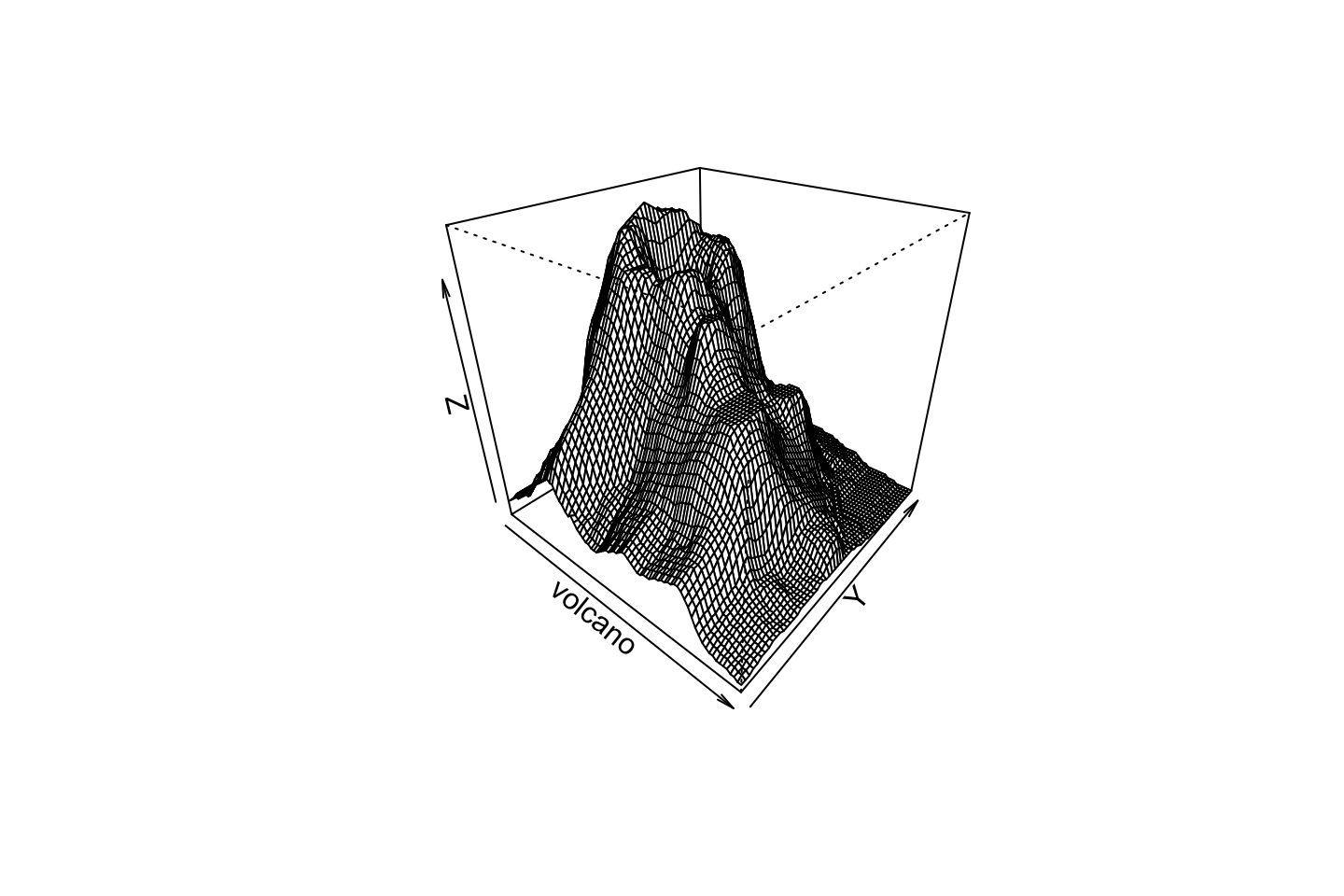
Perspective
persp(volcano, theta=40, phi=30, expand=0.5)
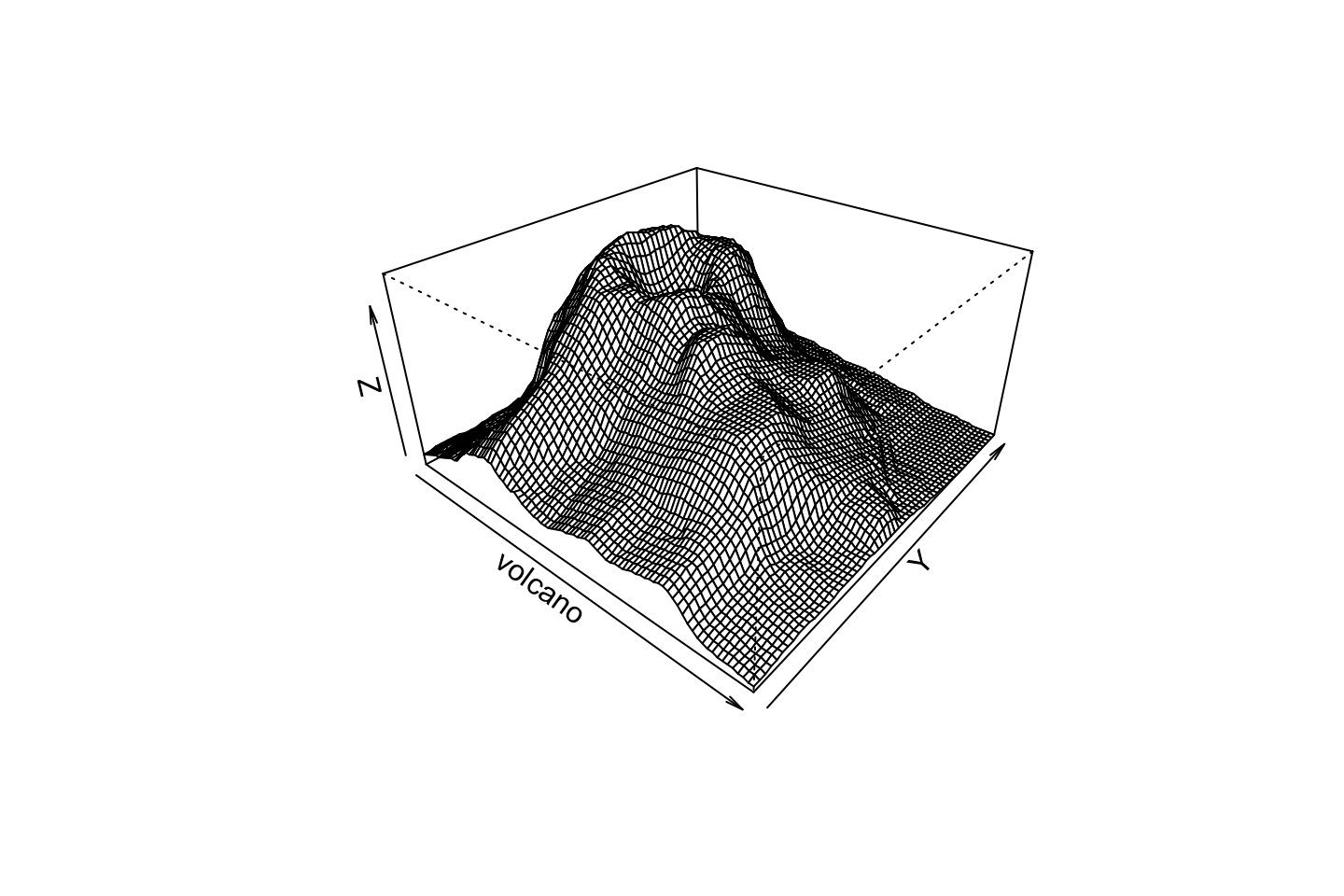
Perspective
persp(volcano, theta=40, phi=30, expand=0.5, col=color[facetcol])
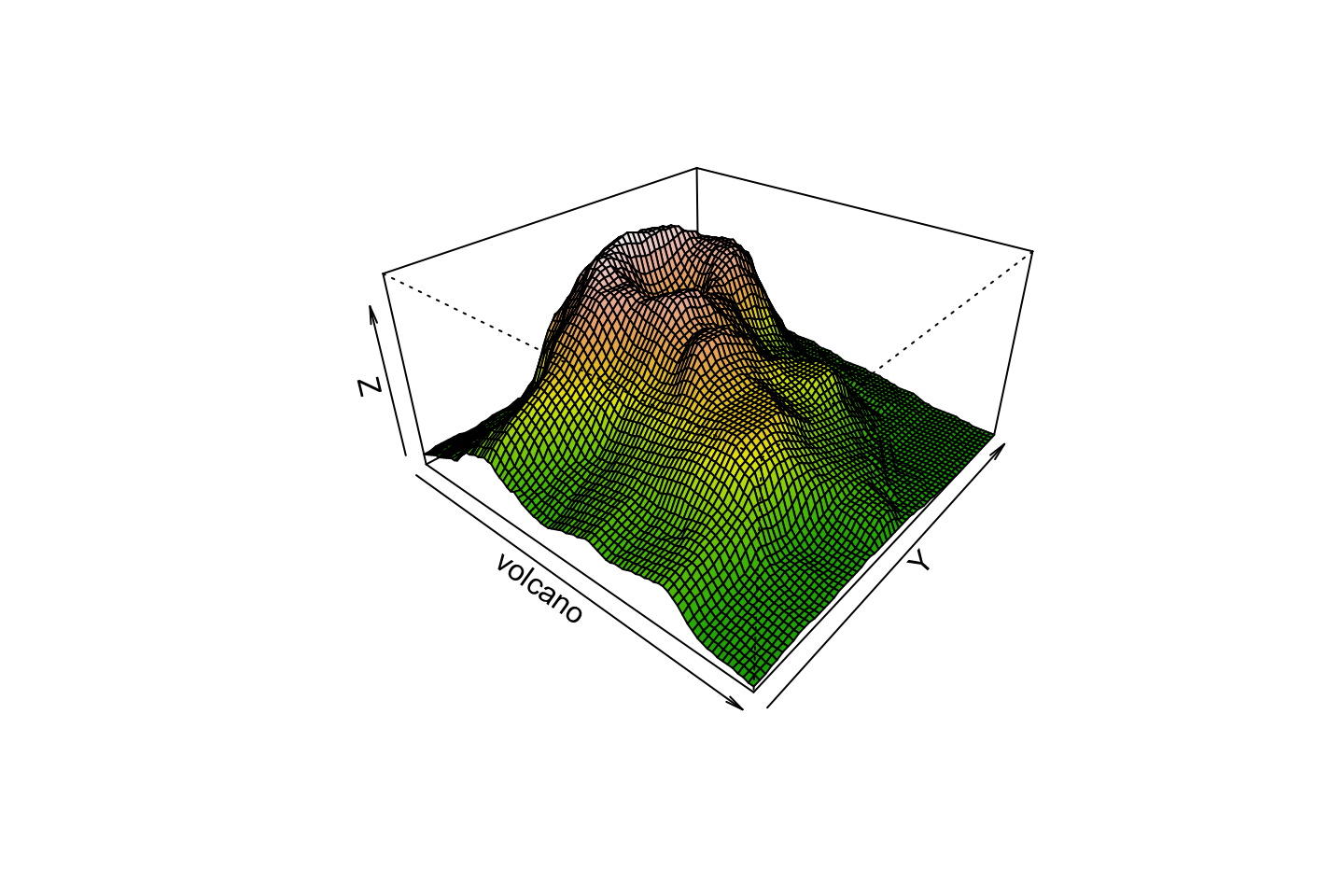
Perspective
persp(volcano, theta=40, phi=30, expand=0.5, col=color[facetcol], border=NA)

Perspective
persp(volcano, theta=40, phi=30, expand=0.5, col=color[facetcol], border=NA,
shade=0.5, ltheta=-40, lphi=40)

Make a movie using animation library
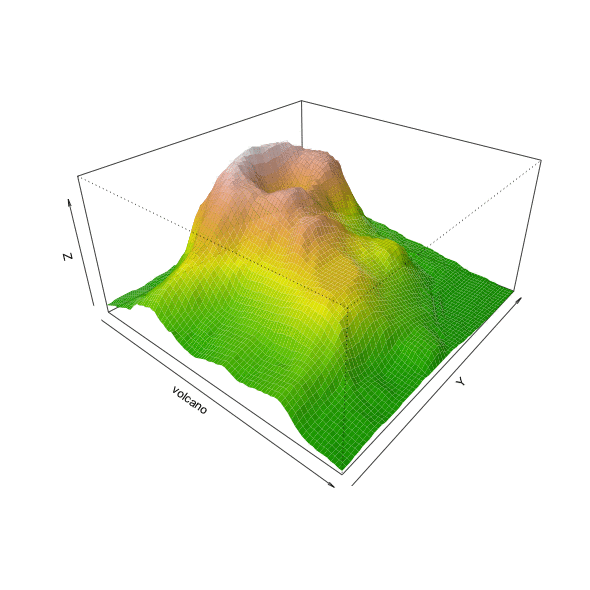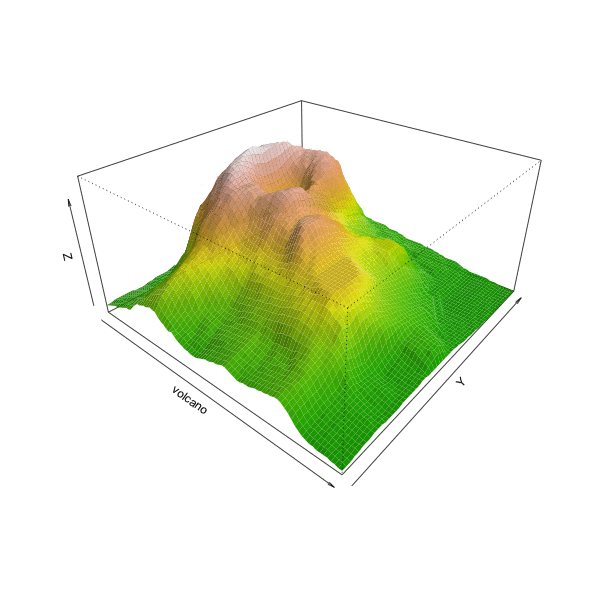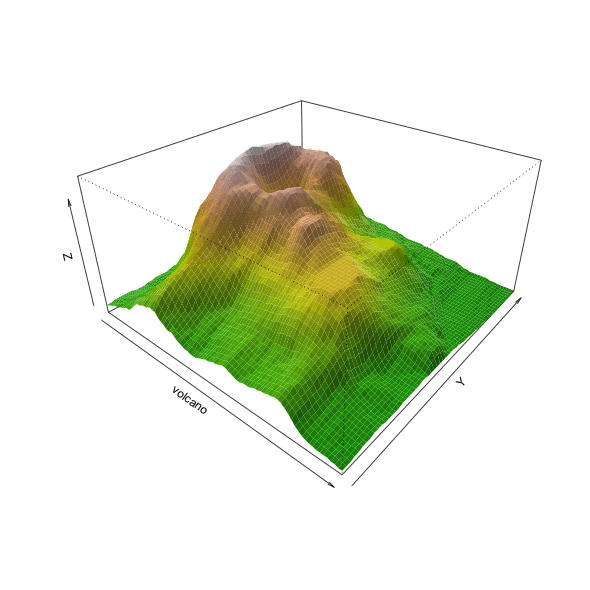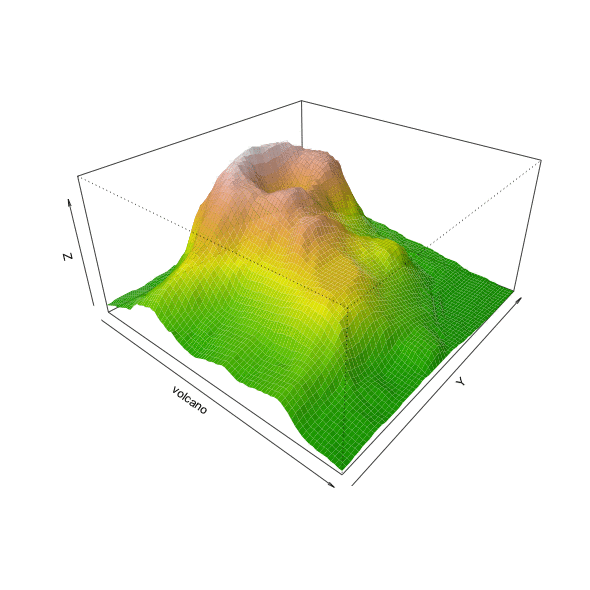
You can animate any parameter
Holiday Homework
Learn Sketch (from MIT) and make a game

Quiz solution
Reading data
lib <- read.delim("libraries.txt", header=TRUE)
The data frame can have any name
Can be different from the file name
I choose to have a short name: lib
First step: what are the variables
The caption says
Read mapping percentages of blood and MBD-enriched fecal samples in three libraries
This means PctReadsMappingBaboon, Type and Library
plot() is not barplot()
plot(PctReadsMappingBaboon ~ ID, data=lib)
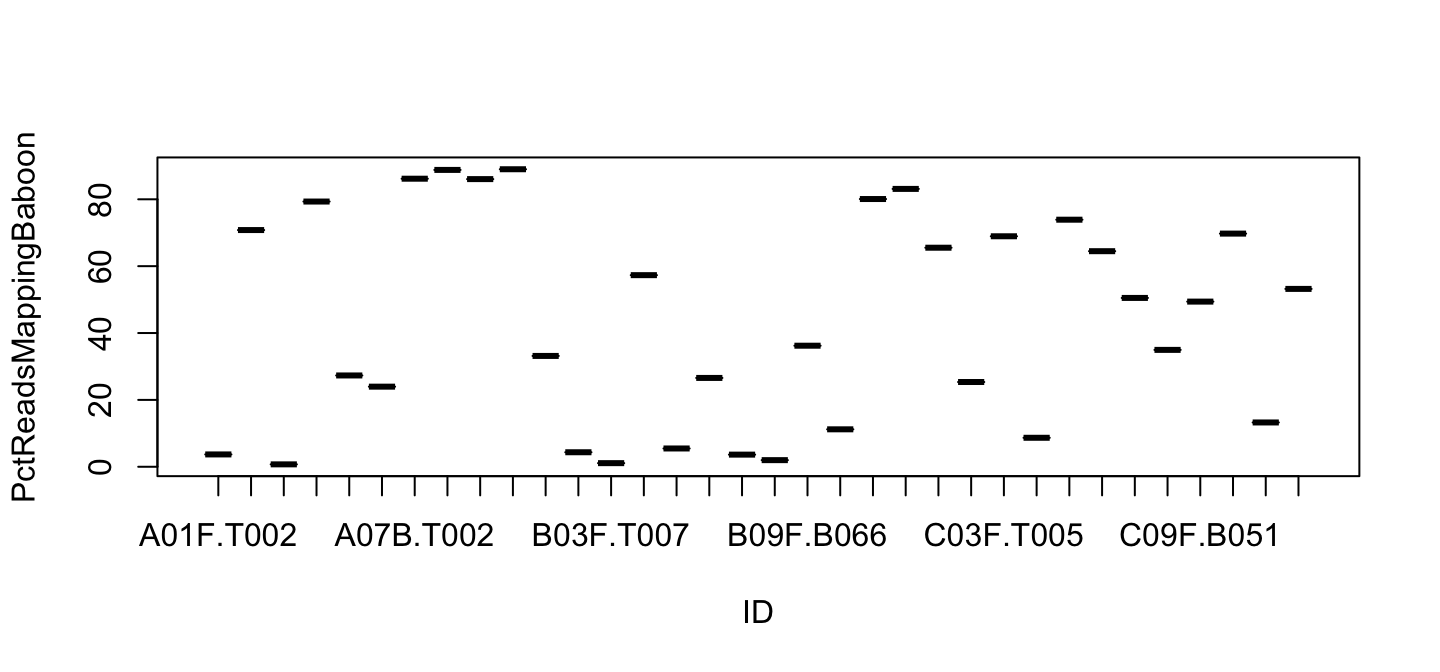
These are not bars. We need barplot
barplot() is not plot()
barplot(PctReadsMappingBaboon ~ ID, data=lib)
Error in barplot.default(PctReadsMappingBaboon ~ ID, data = lib): 'height' must be a vector or a matrix
barplot() does not work wit formulas
It only works with vector, like in the first classes
First plot
barplot(lib$PctReadsMappingBaboon)
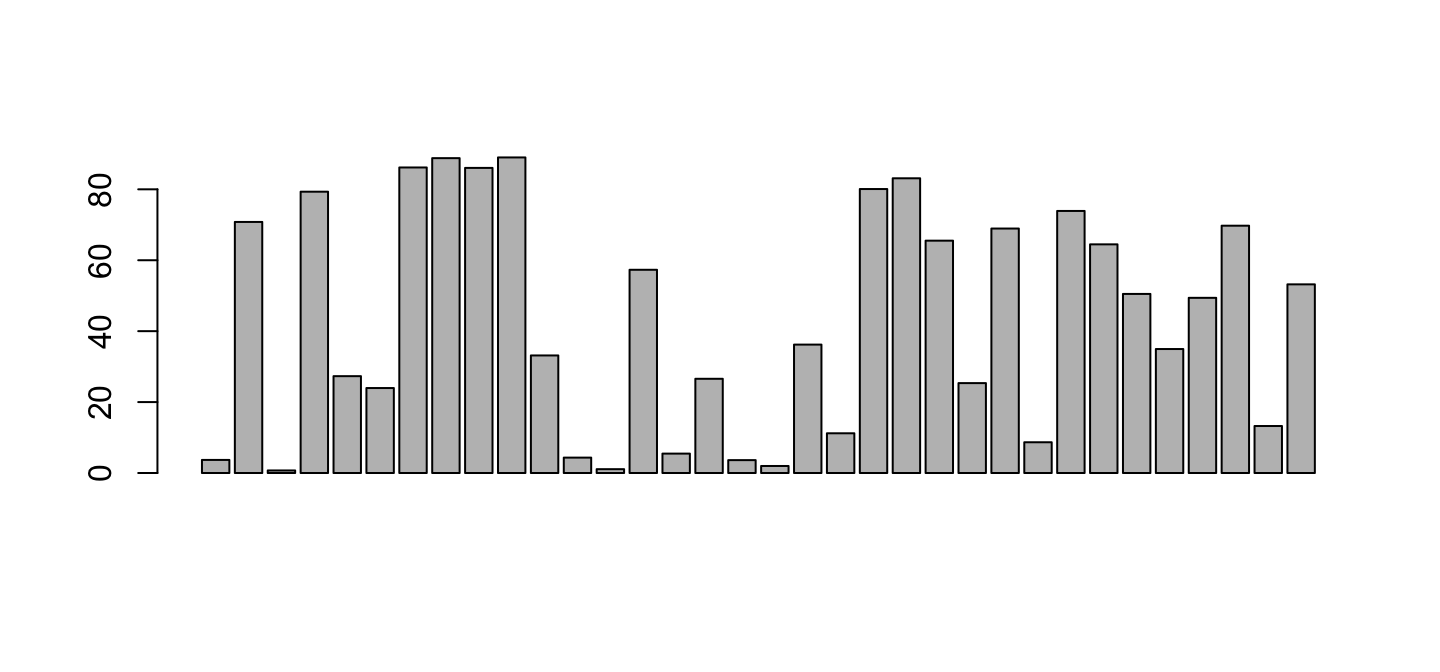 Good. We need to separate Libraries, put names, and paint colors
Good. We need to separate Libraries, put names, and paint colors
Only library A
barplot(lib$PctReadsMappingBaboon[lib$Library=="A"])
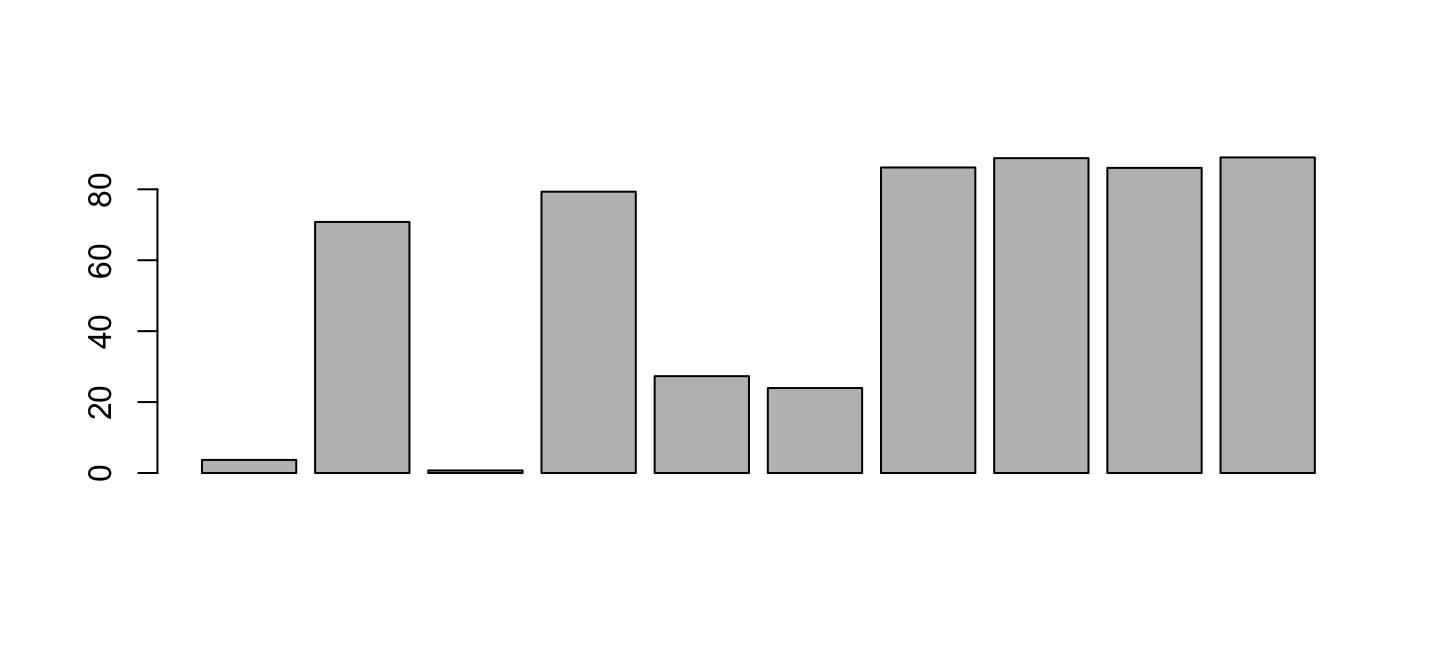 This looks right for the first library
This looks right for the first library
Show also names
barplot(lib$PctReadsMappingBaboon[lib$Library=="A"],
names.arg = lib$ID[lib$Library=="A"])
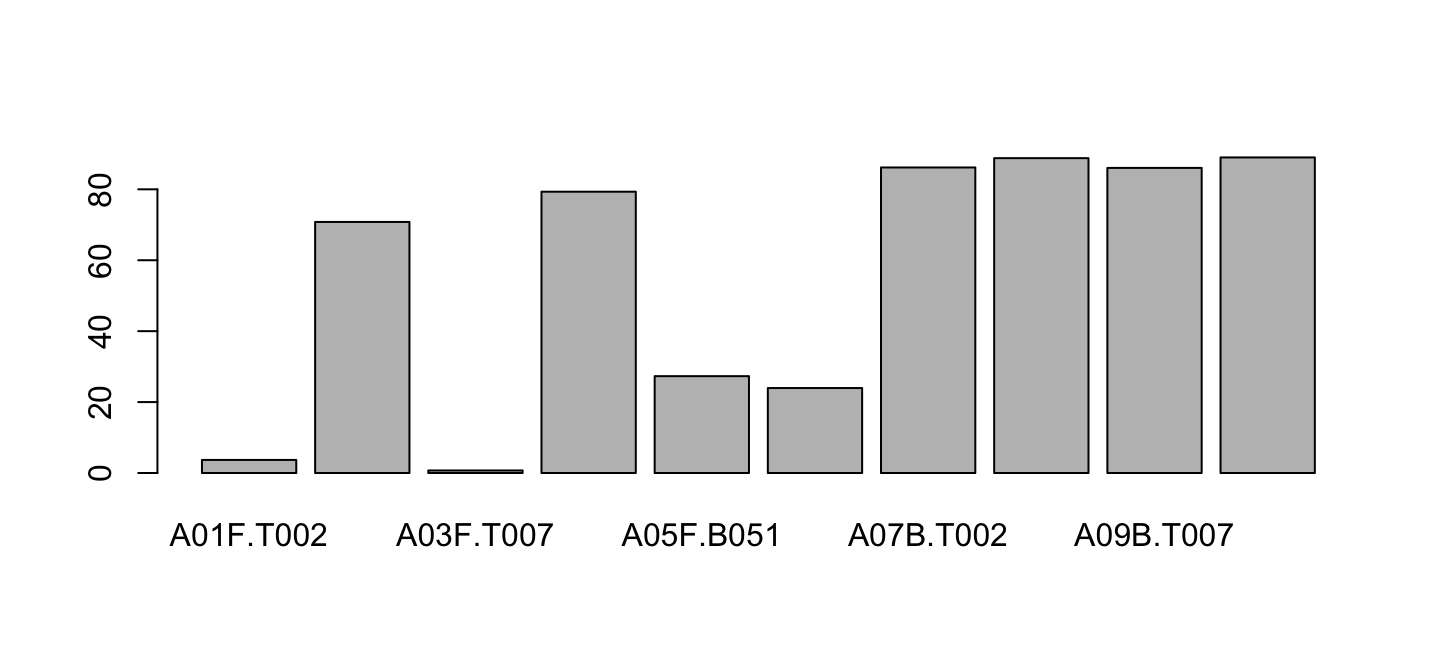
Sideway names
barplot(lib$PctReadsMappingBaboon[lib$Library=="A"],
names.arg = lib$ID[lib$Library=="A"], las=2)
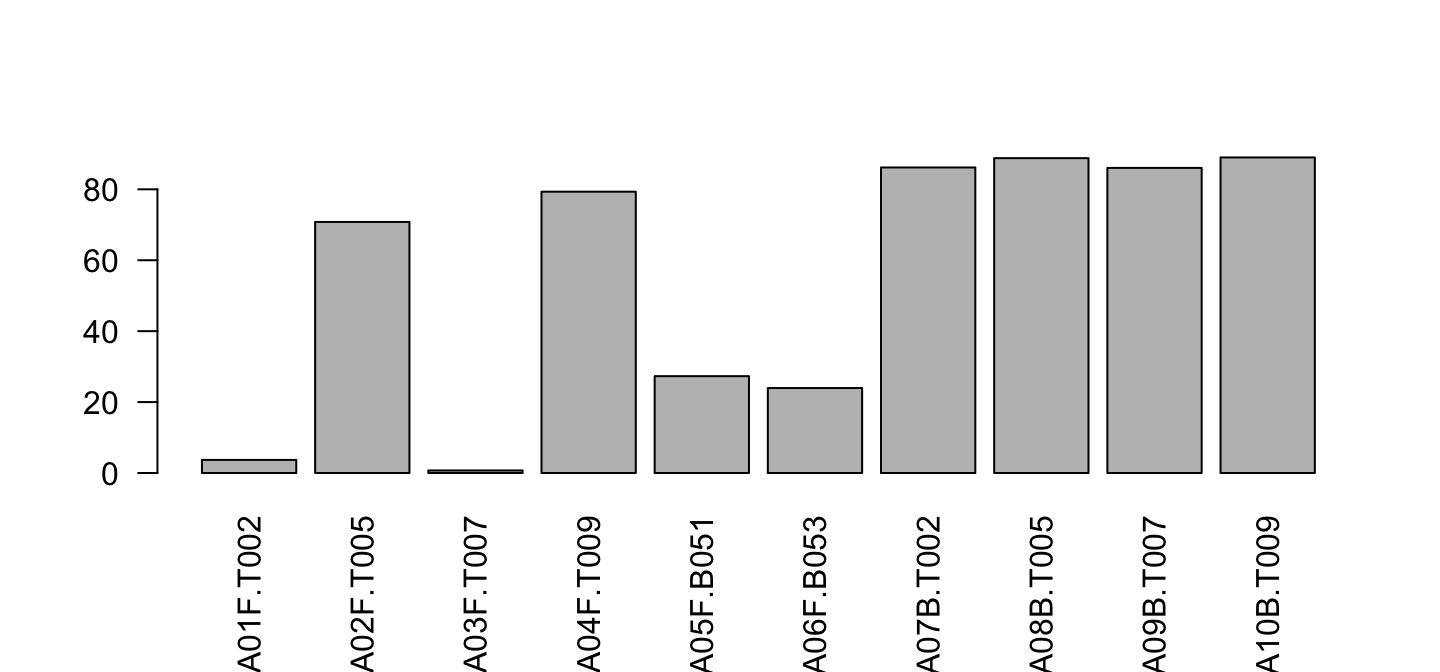
Color depends on Type
barplot(lib$PctReadsMappingBaboon[lib$Library=="A"],
names.arg = lib$ID[lib$Library=="A"], las=2,
col=ifelse(lib$Type[lib$Library=="A"]=="blood",
"red", "brown"))
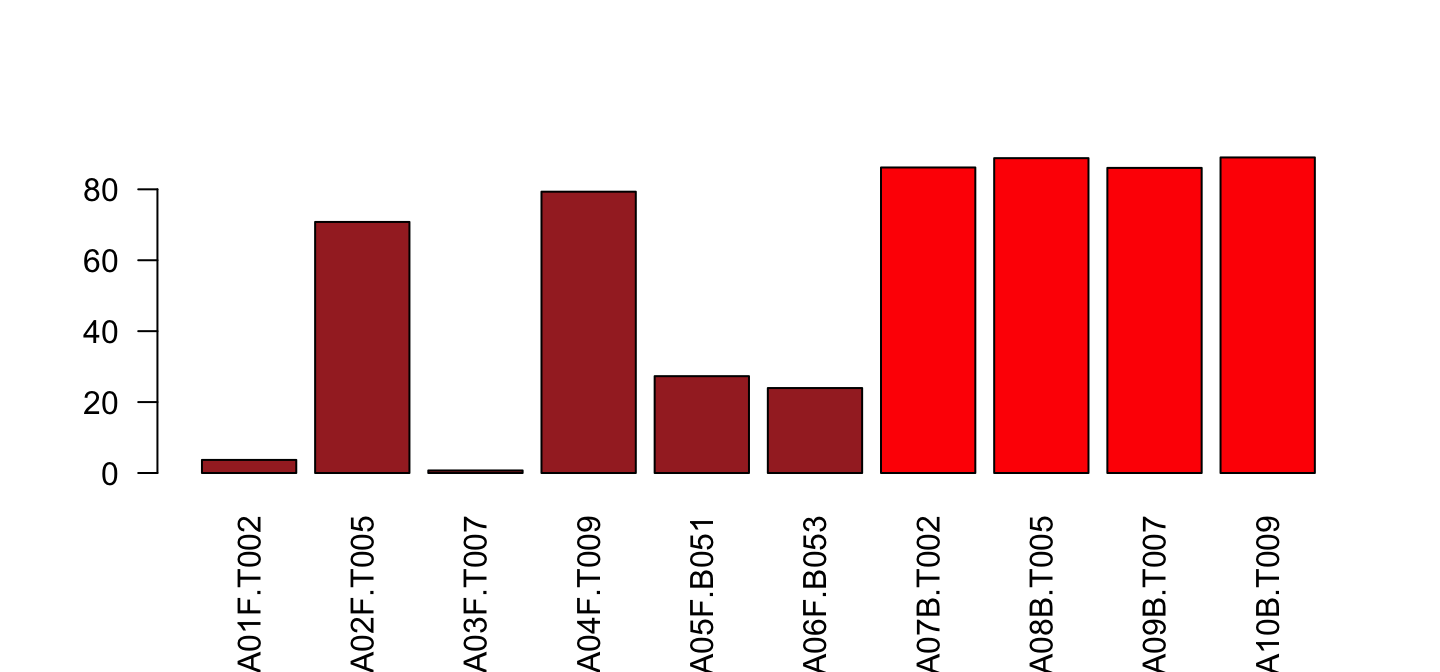
We better choose the library once
part <- lib[lib$Library=="A",]
barplot(part$PctReadsMappingBaboon,
names.arg = part$ID, las=2,
col=ifelse(part$Type=="blood", "red", "#835923"))
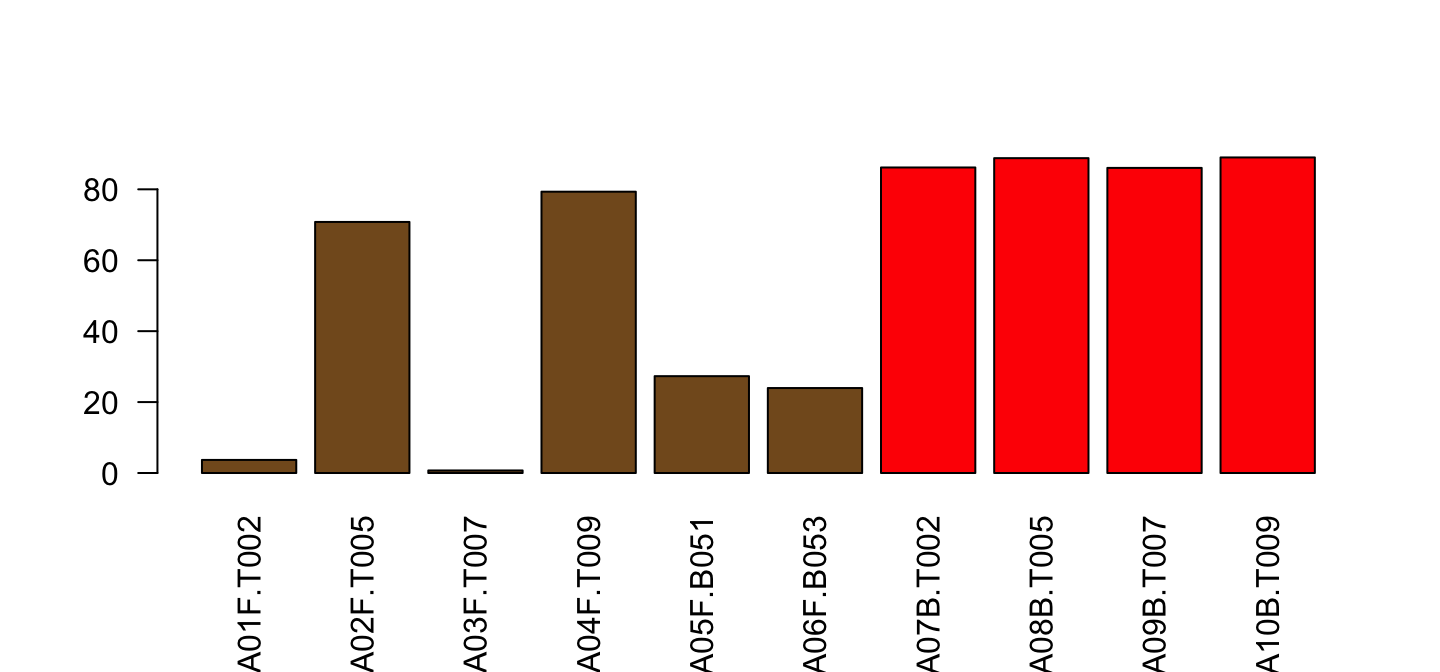
Better separate the library once
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
col=ifelse(part$Type=="blood", "red", "#835923"),
ylim=c(0,100), las=2)
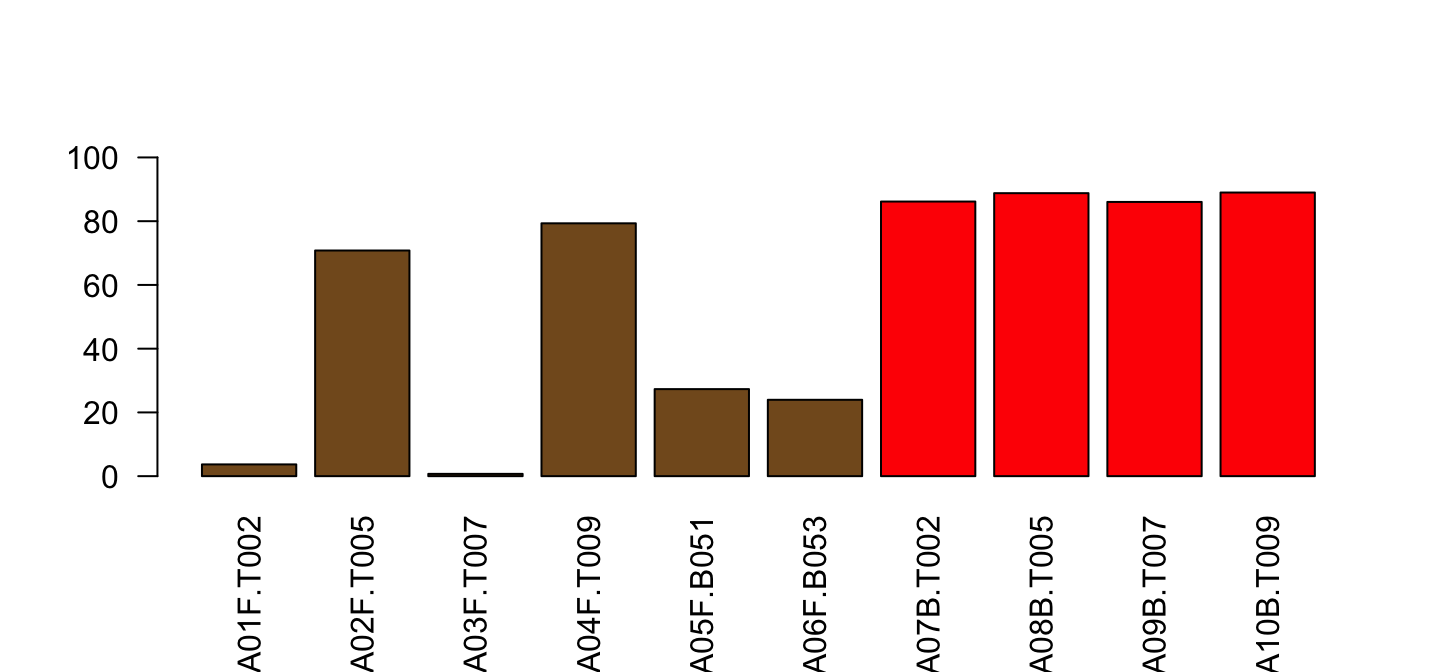
Red line is mean of Pct for Type=="blood"
red_l <- mean(lib$PctReadsMappingBaboon[lib$Type=="blood"]) red_l
[1] 85.52833
Draw the dashed line
part <- lib[lib$Library=="A",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
col=ifelse(part$Type=="blood", "red", "#835923"),
ylim=c(0,100), las=2, main="Library A")
abline(h=red_l, col="red", lty=2, lwd=2)
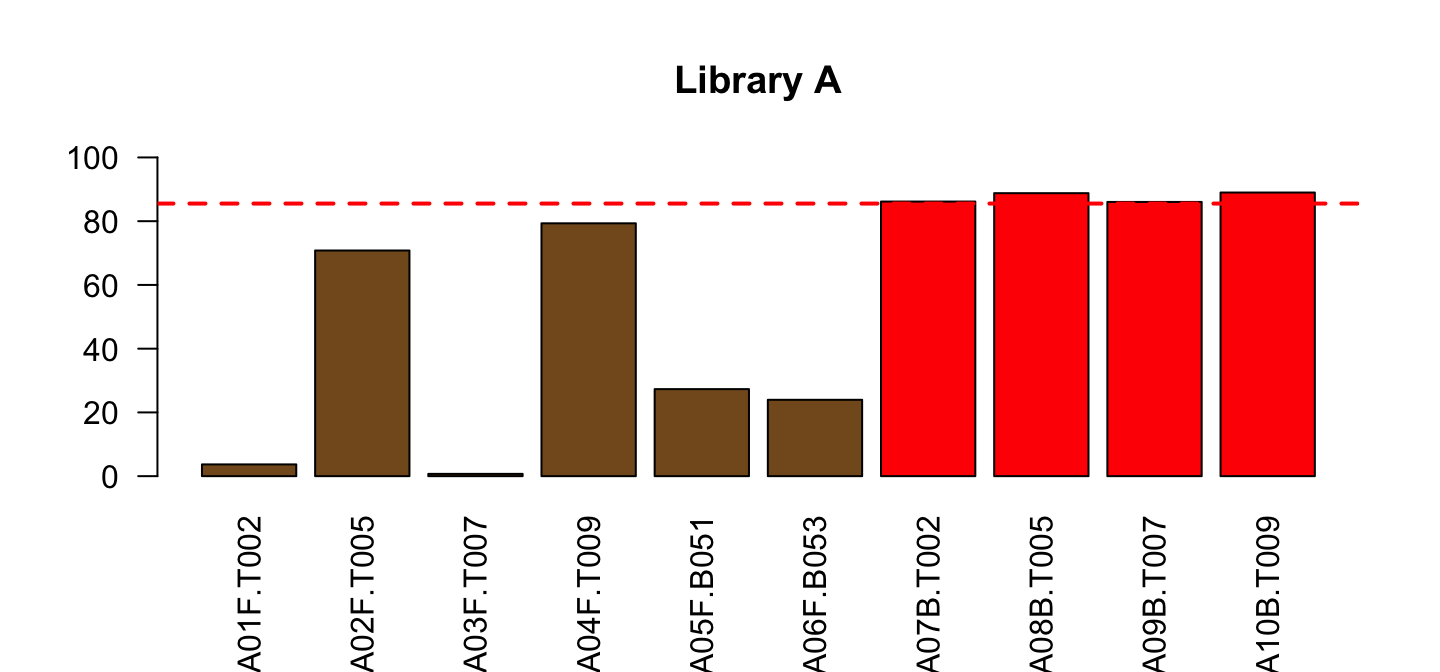
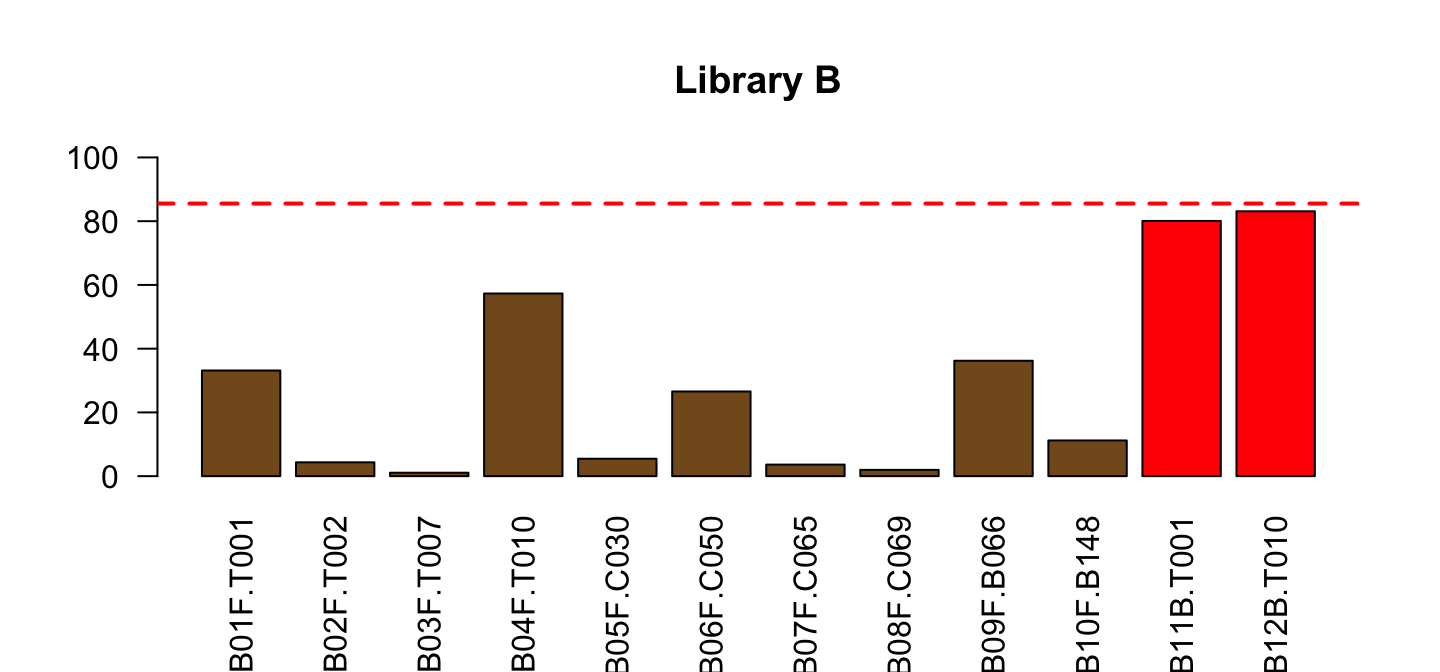
Do the same for Library B
part <- lib[lib$Library=="B",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
col=ifelse(part$Type=="blood", "red", "#835923"),
ylim=c(0,100), las=2, main="Library B")
abline(h=red_l, col="red", lty=2, lwd=2)
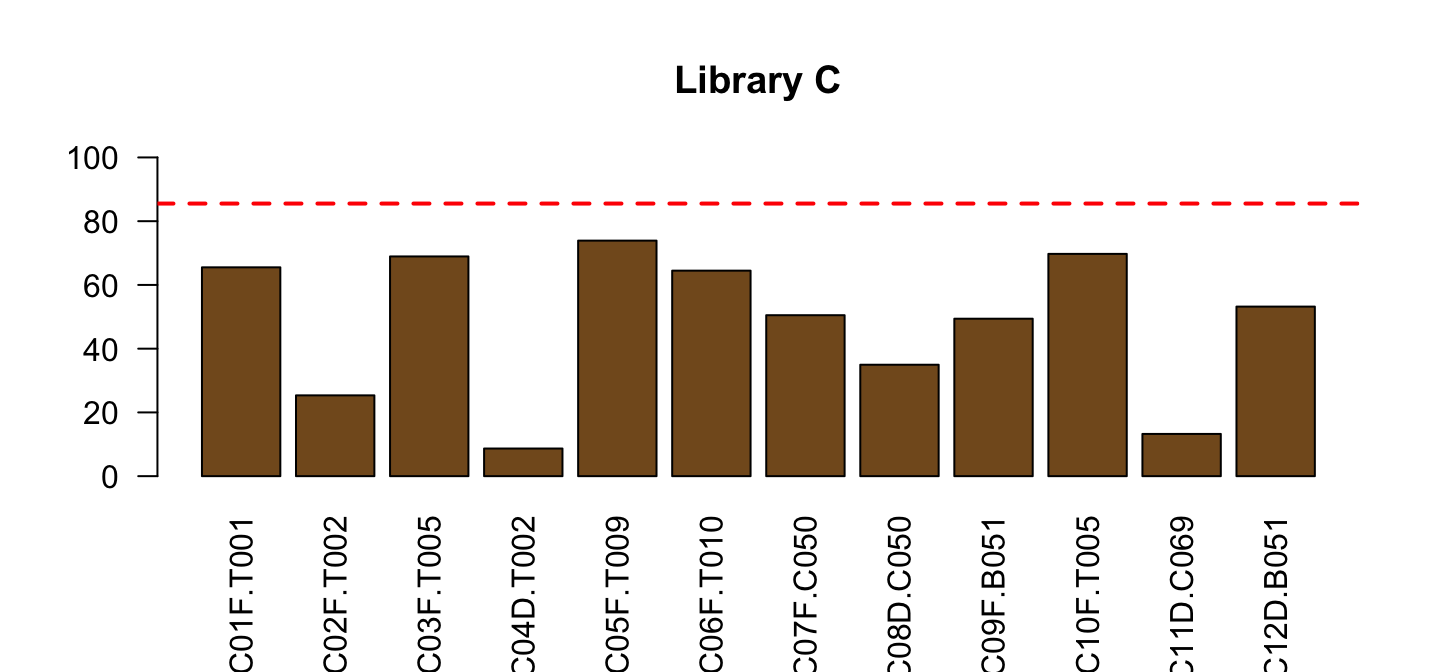
Do the same for Library C
part <- lib[lib$Library=="C",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
col=ifelse(part$Type=="blood", "red", "#835923"),
ylim=c(0,100), las=2, main="Library C")
abline(h=red_l, col="red", lty=2, lwd=2)
All together (see example)
par(mfrow=c(3,1))
part <- lib[lib$Library=="A",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
las=2, ylim=c(0,100),
col=ifelse(part$Type=="blood","red","#835923"))
abline(h=red_l, col="red", lty=2, lwd=2)
part <- lib[lib$Library=="B",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
las=2, ylim=c(0,100),
col=ifelse(part$Type=="blood","red","#835923"))
abline(h=red_l, col="red", lty=2, lwd=2)
part <- lib[lib$Library=="C",]
barplot(part$PctReadsMappingBaboon, names.arg = part$ID,
las=2, ylim=c(0,100),
col=ifelse(part$Type=="blood","red","#835923"))
abline(h=red_l, col="red", lty=2, lwd=2)
par(mfrow=c(1,1))
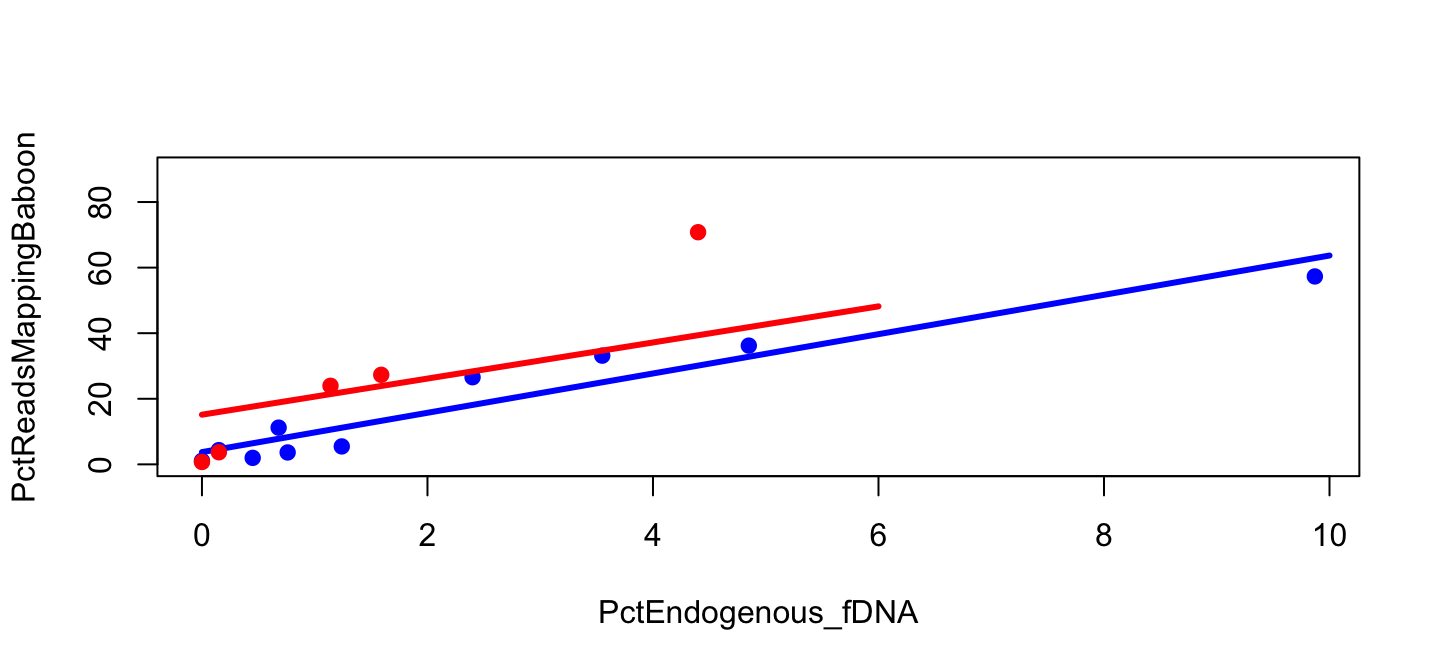
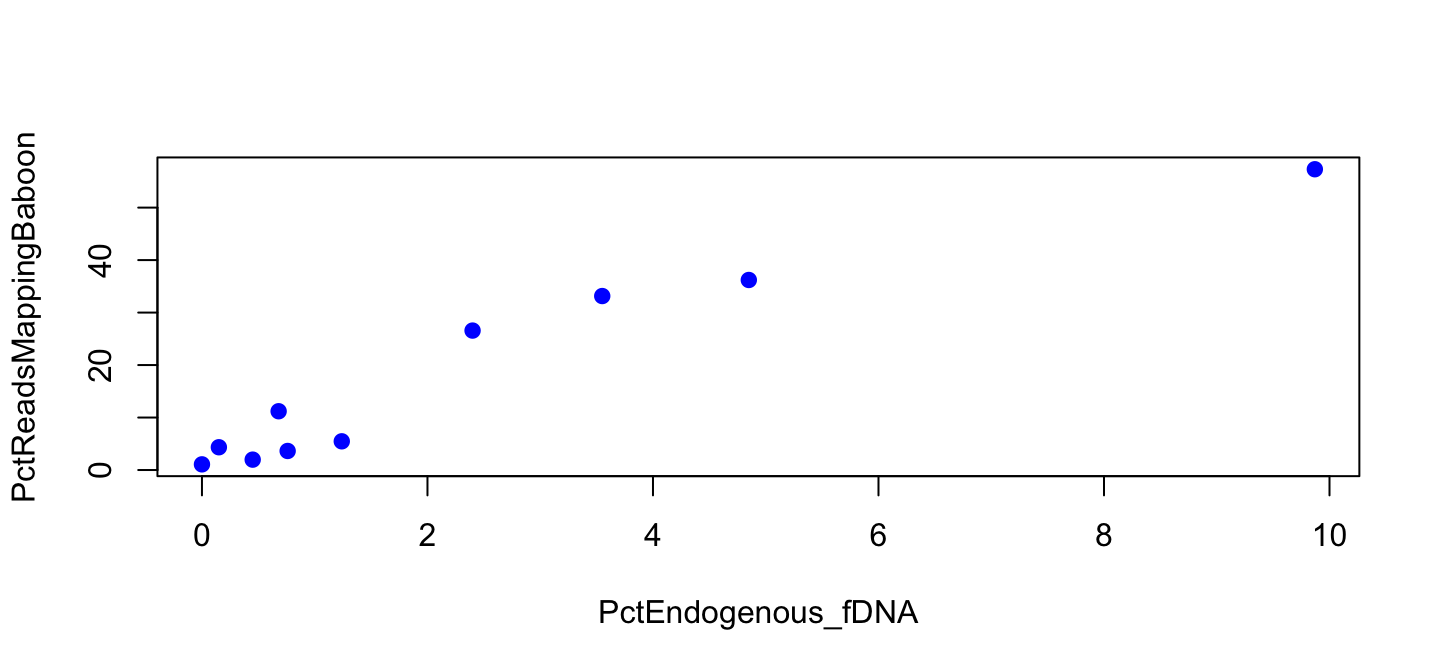
Figure 2
Figure 2 for library B
plot(PctReadsMappingBaboon ~ PctEndogenous_fDNA, data=lib,
subset=Type=="feces" & Library=="B", pch=19, col="blue")
Linear model for the line
model_B <- lm(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="B")
use the same formula, data and subset as in plot()
Predict the line for new data
x_B <- 0:10
y_B <- predict(model_B,
newdata = data.frame(PctEndogenous_fDNA=x_B))
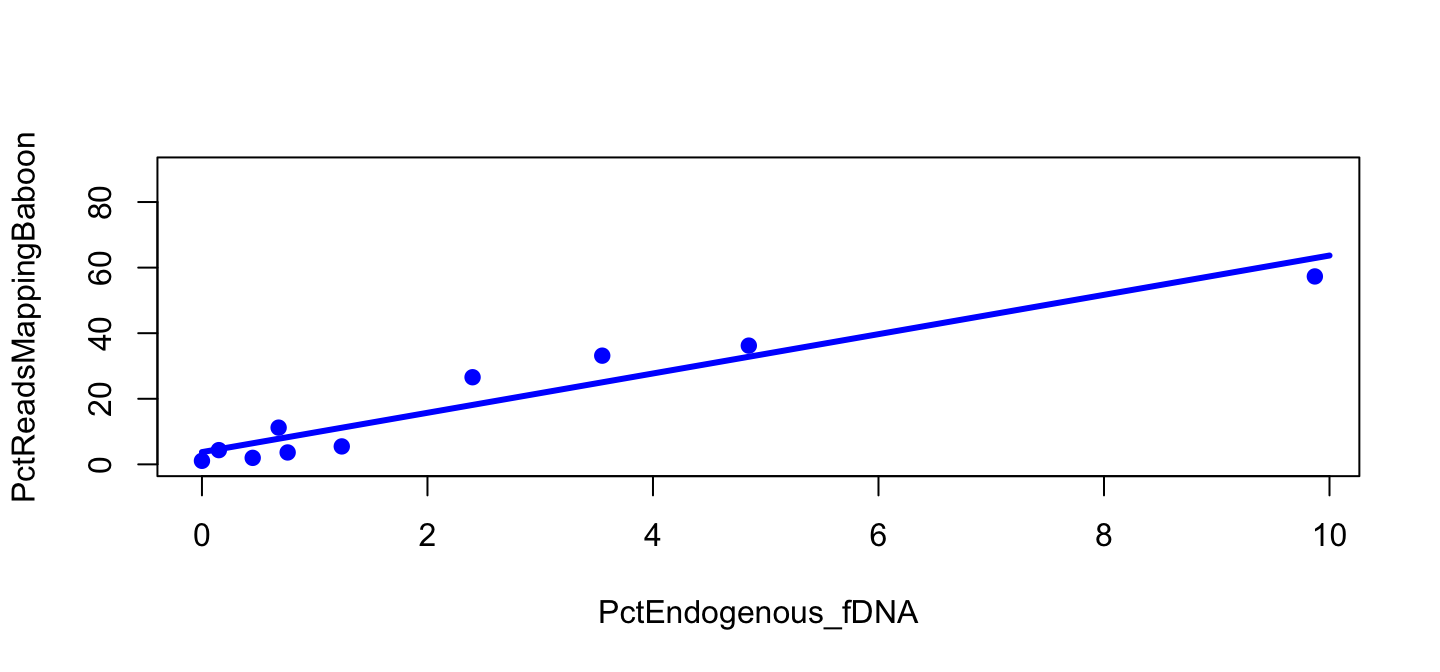
Figure 2 with line
plot(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="B",
pch=19, col="blue", ylim=c(0,90))
lines(y_B ~ x_B, col="blue", lwd=3)
Points for Library A
plot(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="B",
pch=19, col="blue", ylim=c(0,90))
lines(y_B ~ x_B, col="blue", lwd=3)
points(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="A",
pch=19, col="red")
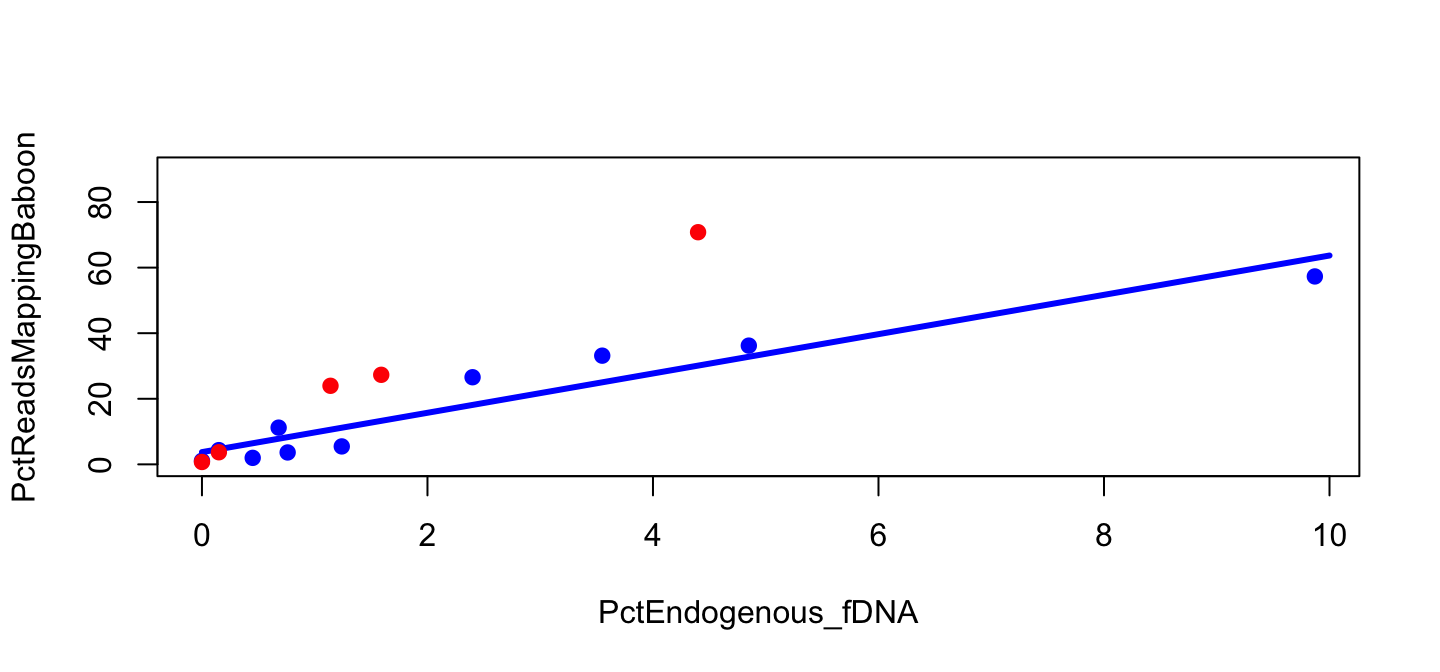
Model for Library A
model_A <- lm(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="A")
x_A <- 0:6
y_A <- predict(model_A,
newdata=data.frame(PctEndogenous_fDNA=x_A))
Finally, plot everything
plot(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="B",
pch=19, col="blue", ylim=c(0,90))
lines(y_B ~ x_B, col="blue", lwd=3)
points(PctReadsMappingBaboon ~ PctEndogenous_fDNA,
data=lib, subset=Type=="feces" & Library=="A",
pch=19, col="red")
lines(y_A ~ x_A, col="red", lwd=3)