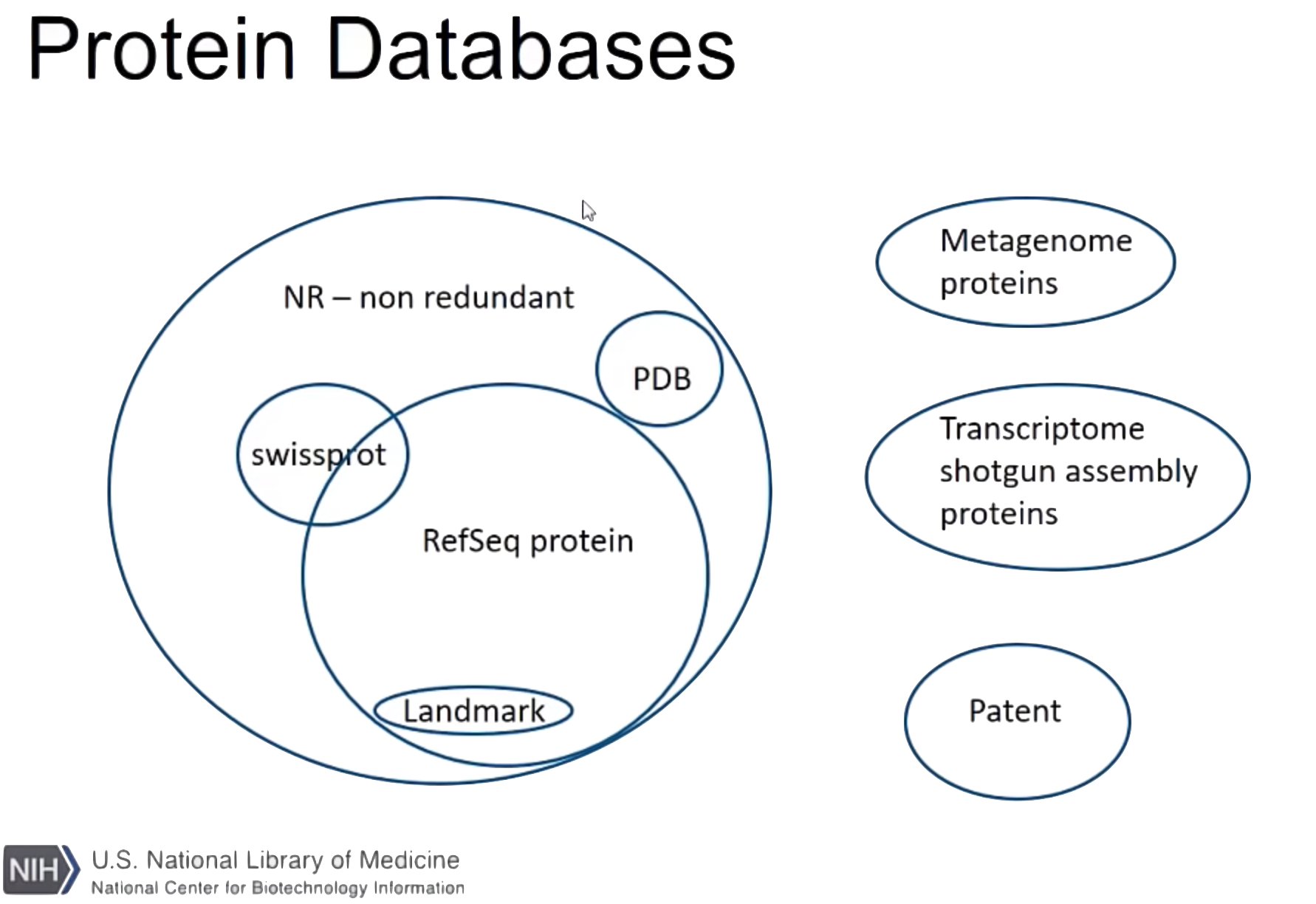
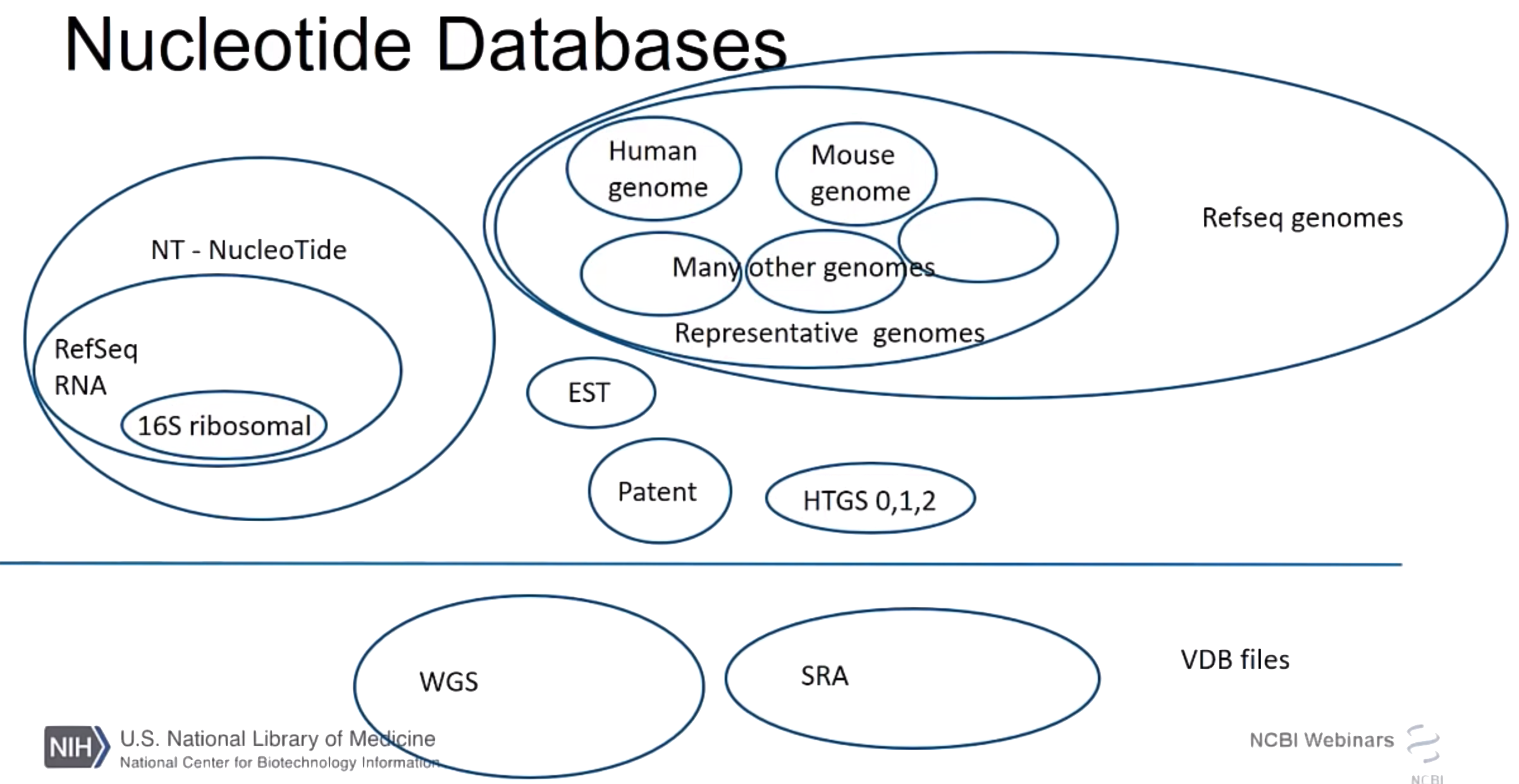
There are two ways of using BLAST
- Going to NCBI’s website: https://blast.ncbi.nlm.nih.gov/
- Runs in NCBI servers with NCBI databases
- Using a command line version
- Runs in our server with our databases
- can also send jobs in NCBI servers with NCBI databases
- Download it from NCBI website
For today we can look at NCBI page