world
September 18th, 2018

world
chile
Population: 17 million
Spanish colony 500 years ago (so language is Spanish)
Independent Republic 200 years ago
First Latin American country to recognize Turkish republic
OECD member, same as Turkey
Everyday life very similar to Turkey

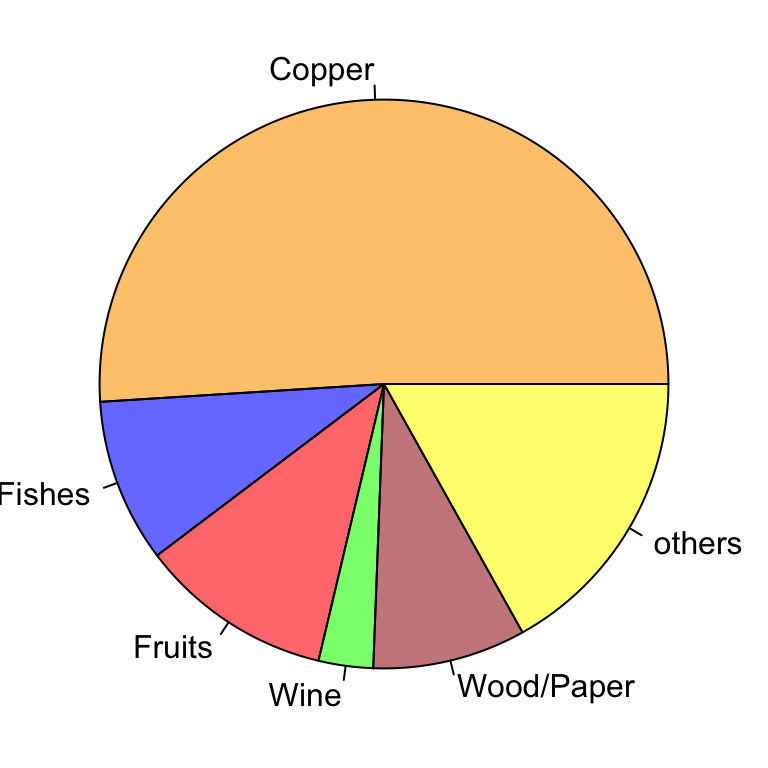
1st world producer of copper
3rd world producer of salmon
Fruits: peaches, grapes, apples, avocado
Wine: exported worldwide
All these industries depend on Biotechnology
Bioleaching is much better that melting copper
The goal is to understand and improve the involved bacteria so this technology can be used extensively
Enables building new mines
It is like discovering petrol reserves for the country
We had a research contract with the main mining company
State owned, big enough to pay for long term research
We focused mainly on 2 questions:
Microorganisms can live in extreme environments
Microorganisms are essential to human life
They are the foundation of all ecosystems
Microorganism live in the most diverse environments.
They are the key to:
But only ~5% of them can be grown in the lab
If we know the DNA sequence for each relevant organism
Ideally the primer probably binds to the organism DNA, and probably do not bind to others organisms.
N.Ehrenfeld, A. Aravena, A. Reyes-Jara, N. Barreto, R. Assar, A. Maass, P. Parada, Design and use of oligonucleotide microarrays for identification of Biomining microorganisms. Advanced Materials Research 71-73 (2009) 155-158.
Since most microbes cannot be isolated, and given current technology, the modern approach is:
Of course the clusters depend on the distance we choose
Most of the sequences from environmental samples are unique
That is, they do not correspond to any known organism
The question is, given a DNA read, which are the “nearest” known organisms?
Here “closest” is in the phylogenetic sense
Currently we use this approach to explore archeological data
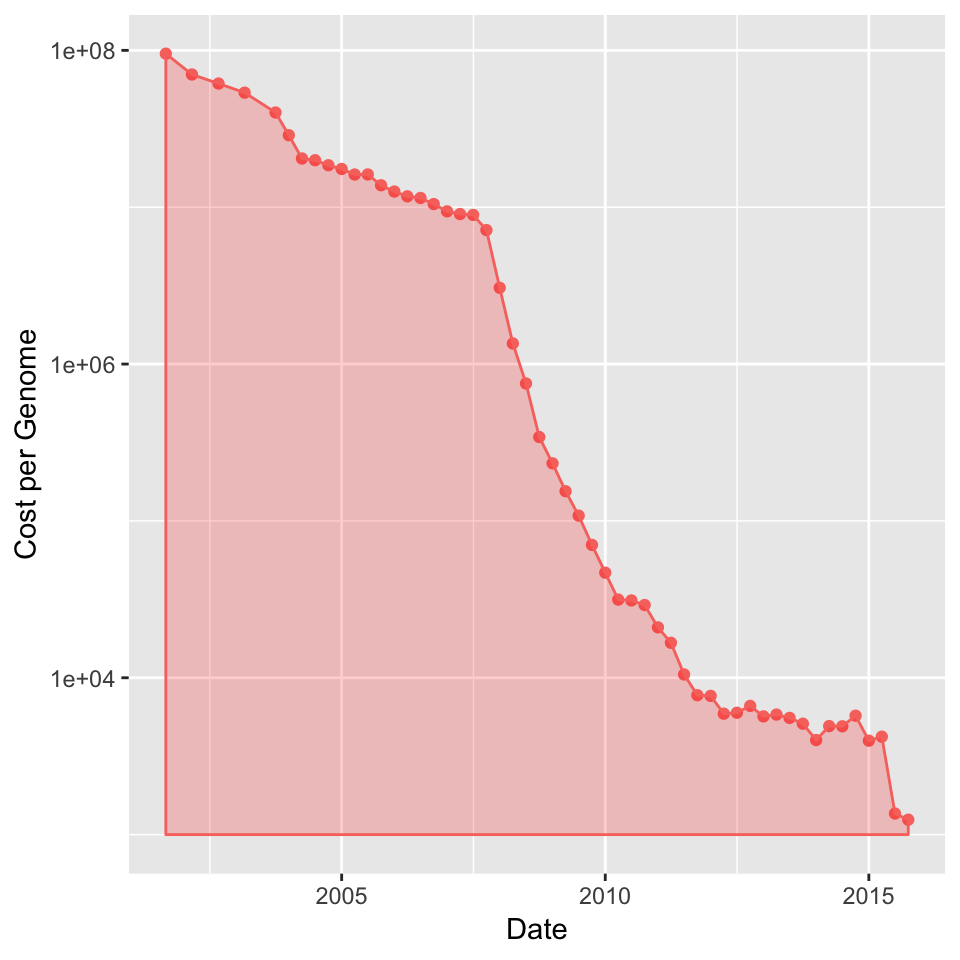
In 2001, the cost of sequencing the first human genome was USD 108
Today you can have your own genome for 1000 USD
The problem is no longer how to do the experiment
Instead is how do we make sense of the results
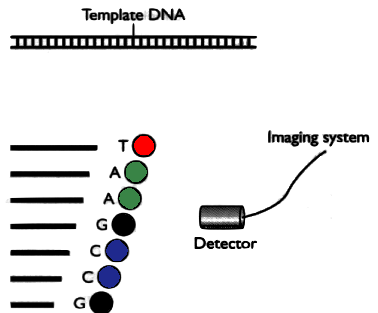
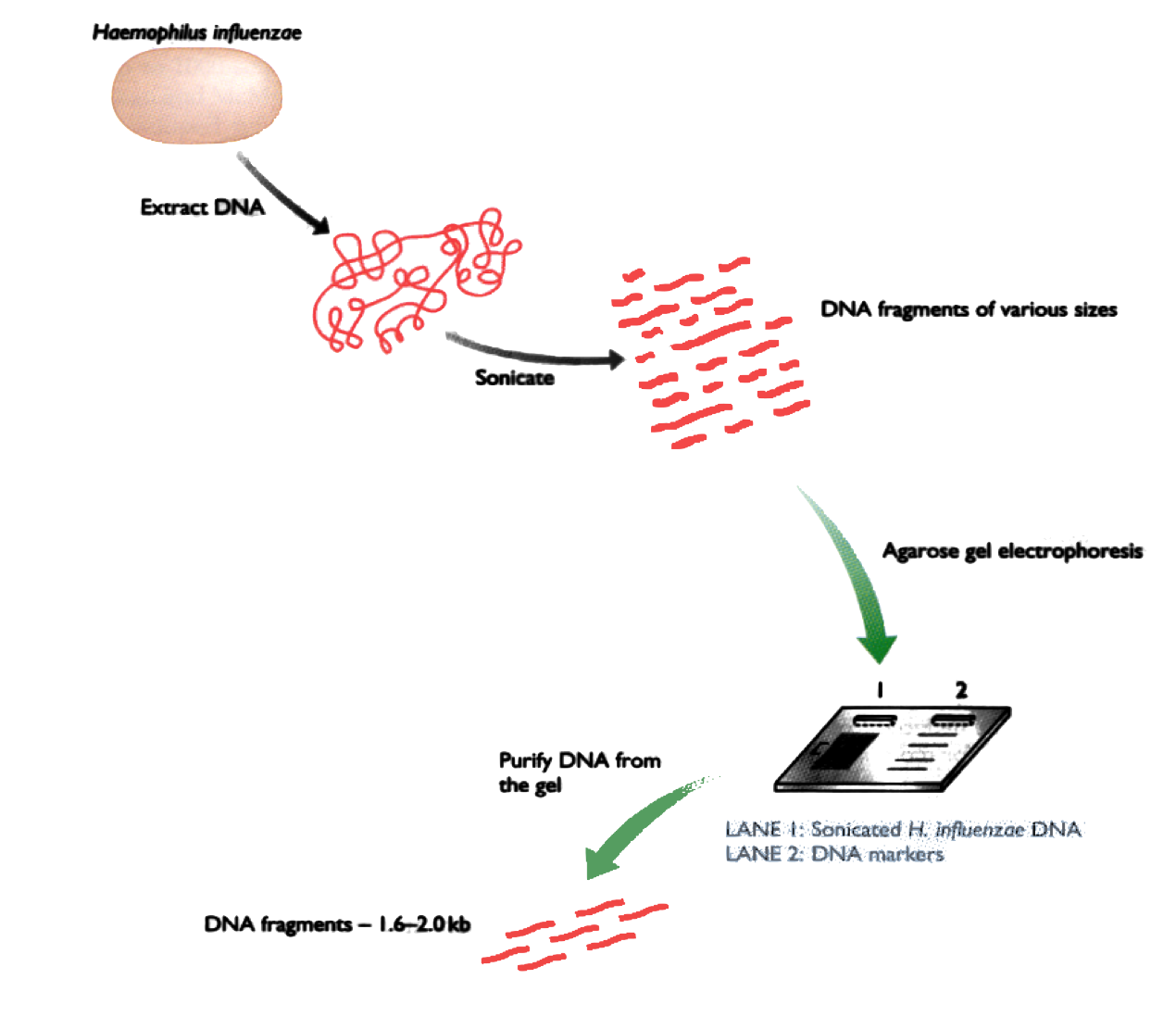
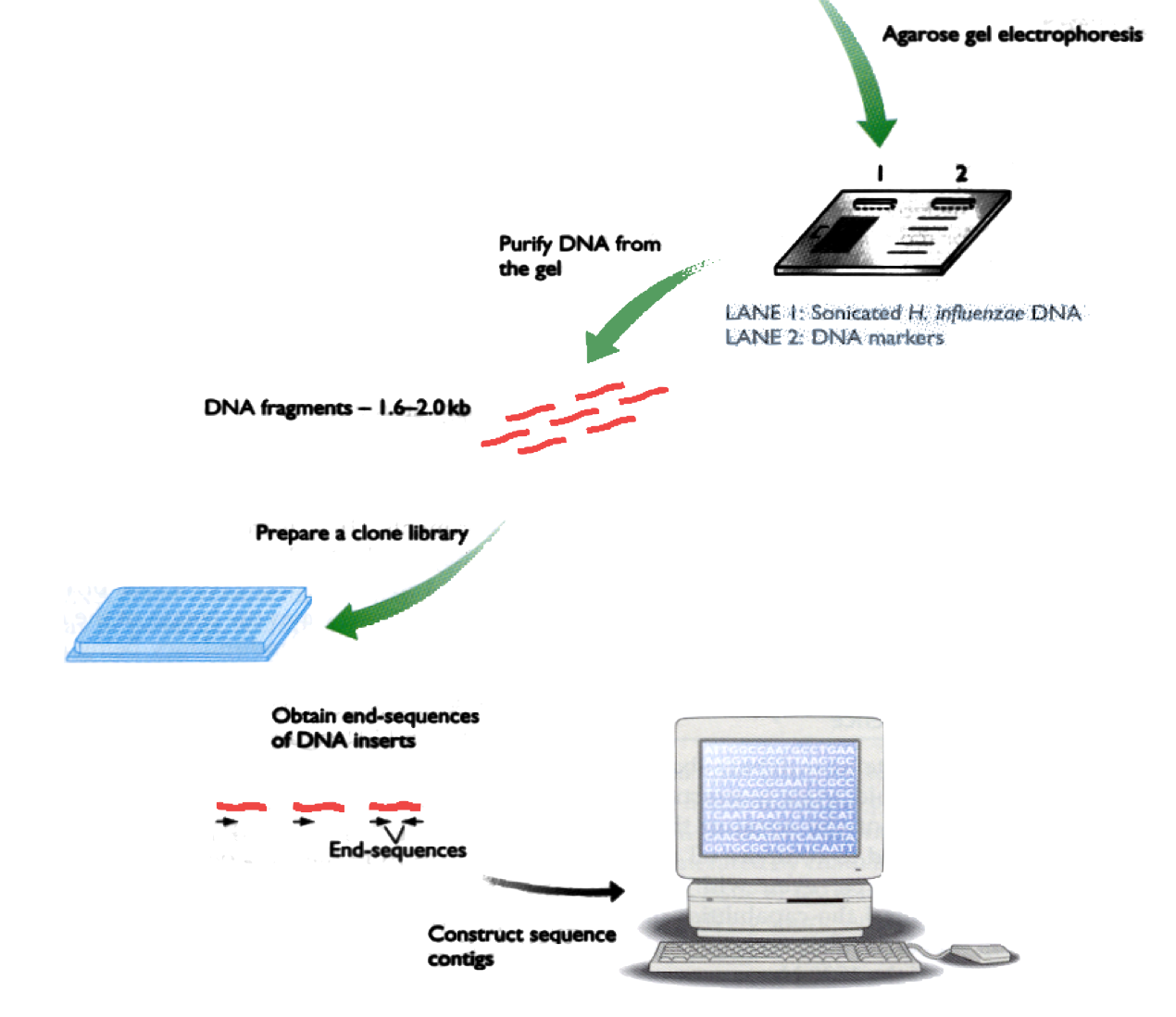
Current technology allows us to read DNA in runs of ~100-600 letters. Imagine a book of 1000 pages:
The problem is to reconstruct the original book