- DNA sequencing
- Pairwise Alignment
- Genome Assembly
- Primer design
- Multiple Alignment
- Finding Binding Sites
- Finding Motifs
- Alignment free methods (composition based)
December 18th, 2017
Big picture
Genomics
Pipelines: putting all together
When we design molecular biology experiments, or when we analyze their results, we need to use several tools in chain
Today we are going to see an example using the NCBBI website
F-Box protein domain

Our challenge is to find proteins in legumes having F-Box and WD-40 domains
Protein domains according to http://pfam.xfam.org/
We need an official definition of each domain
F-box
- motif PF00646.
- alternative motifs (Gupta et al. 2015)
- PF12937, PF13013, PF04300, PF07734, PF07735, PF08268 and PF08387
WD-40
- motif PF00400
Finding proteins with those domains
Using NCBI Conserved Domain Architecture Retrieval Tool (https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi)
Use the query:
[pfam00646,pfam00400]
to look for proteins that contain both domains in the specified order
Results
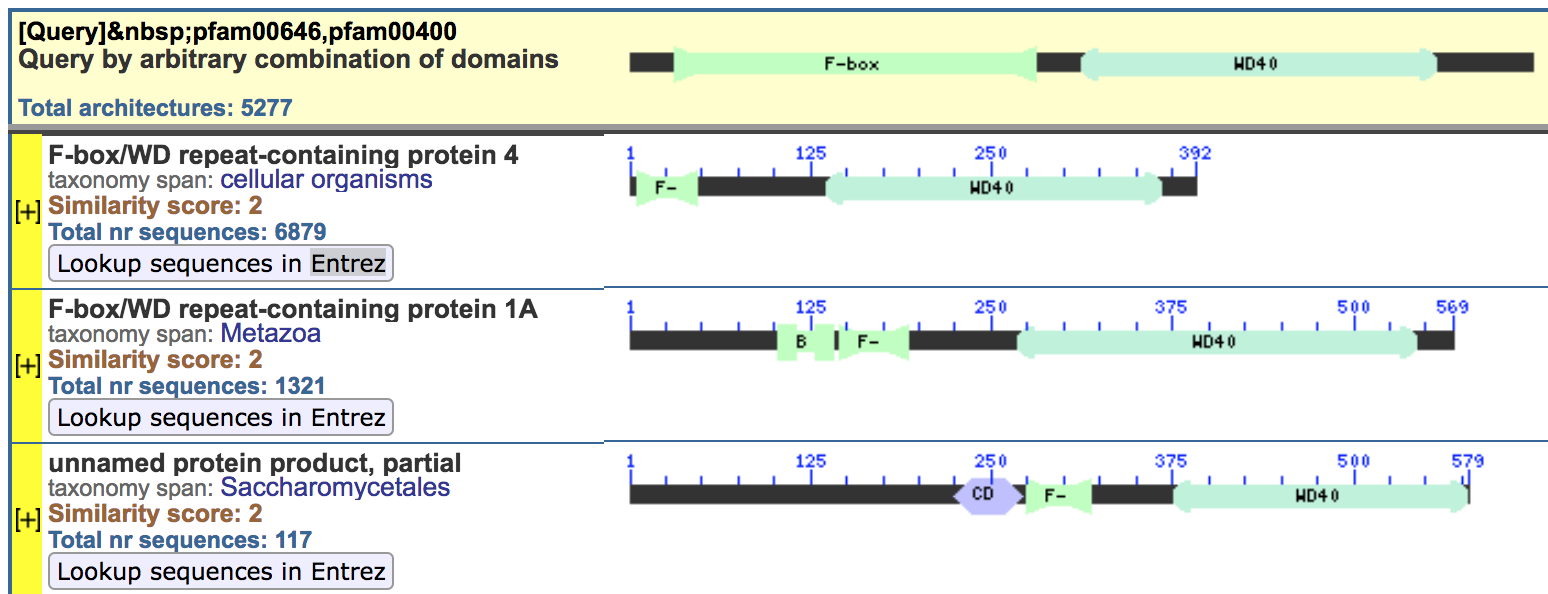
Results
- Several architectures contain both motifs
- Selected the first, with only the relevant domains and nothing else
- Click the button “Lookup sequences in Entrez” to find the list of proteins
Entrez: NCBI website query language
The NCBI website can be searched with many options
The system is called Entrez, like the French for “come in”
It allows us to filter our results using keywords
Filter results: only Legumes
Most of times is a good idea to check the Taxonomy database
Each sequence on GenBank is tagged with a taxon id
Using taxid is more precise than using common names
For example, a protein from human can be labeled “95% similar to mouse”
Is that a human or a mouse protein?
Downloading
For your convenience you can download the sequences
- Decide Format
- Decide Content
In this case we only need accession ids
Finding more proteins
Now we use BLAST to find other proteins in legumes similar to the ones we have
Notice that CDART only has some proteins pre-processed. New sequences take time to be processed
How would you do that?
Save your search strategy
It is essential that your protocol can be replicated
It is a very good idea to save the search strategy in a file
It is also wise to save the output in a text file
Separate by tab or by comma
Process the output
check domains
How many new proteins we find?
Do all of them have the good domains?
Let’s use CDD again, this time looking for motifs on the new proteins \[protein \to\{domains\}\] (the first time was \(domain\to\{proteins\}\))
Next steps
- Keep only proteins with the good domains
- Download the sequences of all proteins
- Download the sequences of the messengers
- Design primers to measure gene expression
- Find literature about gene expression
It is boring to do it one by one
And takes a lot of time
It is easy to make mistakes
It is hard to replicate
Can we do it automatically?
E-tools: Entrez Pipelines
ESearch -> ESummary; ESearch -> EFetch; EPost -> ESummary; EPost -> EFetch; ESearch -> ELink; EPost -> ELink; EPost -> ESearch; ELink -> ESearch; ESearch -> ELink -> ESummary; ESearch -> ELink -> EFetch; EPost -> ESearch -> ESummary; EPost -> ESearch -> EFetch; EPost -> ELink -> ESearch -> ESummary; EPost -> ELink -> ESearch -> EFetch;
Map of E-tools
Use your favorite language
There are Entrez libraries for most languages
For example in R it is called rentrez
Example: analyzing BLAST output
## query subject identity ## 1 gi|255642515|gb|ACU21521.1| gi|356539142|ref|XP_003538059.1| 100.000 ## 2 gi|255642515|gb|ACU21521.1| gi|1150166268|ref|XP_020225874.1| 87.542 ## 3 gi|255642515|gb|ACU21521.1| gi|593697106|ref|XP_007149035.1| 87.629 ## 4 gi|255642515|gb|ACU21521.1| gi|1044577906|ref|XP_017425721.1| 86.254 ## 5 gi|255642515|gb|ACU21521.1| gi|571488796|ref|XP_006591036.1| 88.660 ## 6 gi|255642515|gb|ACU21521.1| gi|950979929|ref|XP_014501961.1| 85.223 ## positives length mismatches gaps q.start q.end s.start s.end evalue ## 1 100.00 291 0 0 1 291 54 344 0 ## 2 92.26 297 31 1 1 291 52 348 0 ## 3 94.16 291 36 0 1 291 50 340 0 ## 4 92.10 291 40 0 1 291 50 340 0 ## 5 88.66 291 0 1 1 291 54 311 0 ## 6 91.07 291 43 0 1 291 50 340 0 ## score q.gi q.ref s.gi s.ref ## 1 612 255642515 ACU21521.1 356539142 XP_003538059.1 ## 2 545 255642515 ACU21521.1 1150166268 XP_020225874.1 ## 3 545 255642515 ACU21521.1 593697106 XP_007149035.1 ## 4 526 255642515 ACU21521.1 1044577906 XP_017425721.1 ## 5 526 255642515 ACU21521.1 571488796 XP_006591036.1 ## 6 522 255642515 ACU21521.1 950979929 XP_014501961.1
Protein ids
These are the genbank ids of all the proteins found
proteins
## [1] "255642515" "1045396645" "1045375294" "1044582125" "1044577908" ## [6] "1044577906" "1021583843" "1021558720" "1012361995" "1012338638" ## [11] "1012260727" "1012202223" "965665445" "965609789" "571507141" ## [16] "571496646" "571488798" "571488796" "356539142" "356501332" ## [21] "950995503" "950979935" "950979929" "950930754" "947065573" ## [26] "357493575" "357458443" "920699279" "920691060" "502169256" ## [31] "502098169" "502090906" "734430373" "734416564" "593701573" ## [36] "593697106" "593562324" "388522749" "1150166268" "1150166270" ## [41] "1117517859" "1012225626" "1150166272" "1117517861" "1012225630" ## [46] "1012225634" "1150128621" "1021534275" "1117375272" "593489431" ## [51] "1150094071" "1044545110" "1117563883" "571553627" "955389649" ## [56] "922350178" "571553636" "922329305" "922350180" "1044556548" ## [61] "1044556546" "1044557823" "1044557821" "1044557819" "951025059" ## [66] "1044557829" "951025065" "1044557825" "1044557827" "951025063" ## [71] "571482571" "571482569" "593689820" "571482575" "571482573" ## [76] "1150095614" "502183121" "1150095616" "828339994" "502183133" ## [81] "502183129" "922400539" "357439909" "828339999" "502183147" ## [86] "502183143" "502183138" "571440442" "571440440" "571440444" ## [91] "356500353" "1117375772" "1117375759" "1117375765" "1117375785" ## [96] "1117375778" "1117546227" "1117546205" "1117375768" "1117375787" ## [101] "1117375775" "1117375781" "1117375790" "955307577" "955307575" ## [106] "1117546230" "1117546224" "955307580" "951017511" "593689824" ## [111] "1117342058" "1117342051" "1117342038" "356537561" "1117342048" ## [116] "1117342055" "1021550847" "1021550849" "1150095590" "1012214348" ## [121] "1044553727" "502103542" "828298200" "1150095620" "951017515" ## [126] "571440447" "593689826" "955384590" "571546627" "950962514" ## [131] "950962510" "356567862" "571546623" "1044534197" "1044534201" ## [136] "357505281" "593441102" "357505277" "593689822" "1150093585" ## [141] "1021530819" "571474527" "1012196136" "1021530815" "1012196130" ## [146] "1021530817" "1012196133" "502132291" "502132289" "571509404" ## [151] "356552535" "1150118542" "1150118544" "502132293" "1044523131" ## [156] "828313695" "1150118546" "356552537" "1150118548" "356503387" ## [161] "571474529" "1012184691" "1012184695" "357509397" "1150136062" ## [166] "955314277" "593562195" "1044582959" "950983855" "356553464" ## [171] "356499483" "1150127909" "828296784" "357495283" "1117372388" ## [176] "1012199286" "1021533997"
What are their domains?
domains <- entrez_link(dbfrom = "protein", id=proteins, by_id=TRUE, db="cdd") doms <- lapply(domains, function(m) m$links$protein_cdd_concise_2) udomains <- unique(unlist(doms)) udomains
## [1] "330360" "315592" "306992" "238121" "225201" "321319" "312123" ## [8] "294672" "312941" "330537"
But what do these id mean?
dom.summary <- entrez_summary("cdd", udomains)
dom.title <- extract_from_esummary(dom.summary, "title")
prot.domains <- sapply(doms, function(d) paste(dom.title[d], collapse=" "))
prot.domains
## [1] "WD40 F-box-like" "WD40 F-box" ## [3] "WD40 F-box-like" "WD40 F-box-like" ## [5] "F-box-like WD40" "WD40 F-box-like" ## [7] "WD40 F-box" "WD40 F-box-like" ## [9] "WD40 F-box-like" "WD40 F-box-like" ## [11] "WD40 F-box" "WD40 F-box-like" ## [13] "WD40 F-box-like" "WD40 F-box" ## [15] "WD40 F-box" "WD40" ## [17] "WD40 F-box-like" "WD40 F-box-like WD40" ## [19] "WD40 F-box-like" "WD40 F-box" ## [21] "WD40 F-box-like" "" ## [23] "WD40 F-box-like" "WD40 F-box" ## [25] "WD40 F-box" "WD40 F-box" ## [27] "WD40 F-box-like" "WD40 F-box-like" ## [29] "WD40 F-box" "WD40 F-box-like" ## [31] "WD40 F-box" "WD40" ## [33] "WD40 F-box" "WD40 F-box" ## [35] "WD40 F-box-like" "WD40 F-box-like" ## [37] "WD40" "F-box-like WD40" ## [39] "WD40 F-box" "WD40 F-box" ## [41] "WD40 F-box-like" "WD40" ## [43] "WD40 F-box" "WD40 F-box-like" ## [45] "WD40" "WD40" ## [47] "WD40 F-box-like" "" ## [49] "WD40 F-box" "SGNH_hydrolase WD40 F-box-like" ## [51] "WD40 F-box-like" "WD40" ## [53] "WD40 F-box" "WD40" ## [55] "WD40" "" ## [57] "WD40" "" ## [59] "" "WD40" ## [61] "WD40" "WD40 LisH Dyp_perox" ## [63] "WD40 LisH Dyp_perox" "WD40 LisH Dyp_perox" ## [65] "WD40 LisH Dyp_perox" "WD40 LisH Dyp_perox" ## [67] "WD40 LisH Dyp_perox" "WD40 LisH" ## [69] "WD40 LisH" "WD40 LisH" ## [71] "WD40 LisH Dyp_perox" "WD40 LisH Dyp_perox" ## [73] "WD40 LisH Dyp_perox" "WD40 LisH" ## [75] "WD40 LisH" "WD40 LisH" ## [77] "WD40 Med15 LisH" "WD40 LisH" ## [79] "WD40 LisH Med15" "WD40 LisH" ## [81] "WD40 LisH Med15" "WD40 LisH" ## [83] "WD40 LisH" "WD40 Med15 LisH" ## [85] "WD40 LisH" "WD40 LisH Med15" ## [87] "WD40 Med15 LisH" "WD40 LisH Dyp_perox" ## [89] "WD40 LisH Dyp_perox" "WD40 LisH" ## [91] "WD40 LisH" "WD40 LisH" ## [93] "WD40 LisH" "WD40 LisH" ## [95] "WD40 LisH" "WD40 LisH" ## [97] "WD40 LisH" "WD40 LisH" ## [99] "WD40 LisH" "WD40 LisH" ## [101] "WD40 LisH" "WD40 LisH" ## [103] "WD40 LisH" "WD40 LisH Dyp_perox" ## [105] "WD40 LisH Dyp_perox" "WD40 LisH" ## [107] "WD40 LisH" "WD40 LisH" ## [109] "F-box" "F-box" ## [111] "WD40" "WD40 LisH" ## [113] "WD40 LisH" "F-box" ## [115] "WD40 LisH" "WD40 LisH" ## [117] "WD40 LisH" "WD40 LisH" ## [119] "F-box" "" ## [121] "F-box" "WD40 LisH" ## [123] "WD40 LisH" "F-box" ## [125] "" "WD40 LisH Dyp_perox" ## [127] "F-box" "WD40 LisH" ## [129] "WD40 LisH" "WD40 LisH Med15" ## [131] "WD40 LisH" "WD40 LisH" ## [133] "WD40 LisH" "WD40 LisH" ## [135] "WD40 LisH" "WD40 LisH" ## [137] "WD40 LisH" "WD40 LisH" ## [139] "F-box" "WD40 LisH Med15" ## [141] "WD40" "WD40 LisH" ## [143] "" "WD40 Med15 LisH" ## [145] "WD40 LisH" "WD40 LisH" ## [147] "WD40 LisH" "WD40 LisH" ## [149] "WD40 LisH" "WD40 Amelogenin LisH" ## [151] "WD40 Amelogenin LisH" "WD40 LisH" ## [153] "WD40 LisH" "WD40 LisH" ## [155] "WD40" "WD40 LisH" ## [157] "WD40 LisH" "WD40 Amelogenin LisH" ## [159] "WD40 LisH" "F-box" ## [161] "WD40 LisH" "F-box" ## [163] "F-box" "F-box" ## [165] "F-box" "F-box" ## [167] "WD40" "WD40" ## [169] "WD40" "WD40" ## [171] "WD40" "WD40" ## [173] "" "" ## [175] "" "" ## [177] ""
Keep only proteins that have both domains
wd40.id <- names(dom.title)[dom.title=="WD40"]
fbox.id <- names(dom.title)[dom.title %in% c("F-box", "F-box-like")]
has.wd40 <- sapply(doms, function(d) length(intersect(d, wd40.id))>0)
has.fbox <- sapply(doms, function(d) length(intersect(d, fbox.id))>0)
has.both <- has.fbox & has.wd40
table(has.both)
## has.both ## FALSE TRUE ## 133 44
Download the protein sequences
prot.seq <- entrez_fetch(db="protein", rettype = "fasta", id=proteins[has.both]) write(prot.seq, file="ncbi-proteins-wd40-fbox.faa")
Find the messengers that code for the proteins
messn <- entrez_link(dbfrom = "protein", id=proteins, by_id=TRUE,
linkname="protein_nuccore_mrna")
messn <- sapply(messn, function(m) m$links$protein_nuccore_mrna)
messn.seq <- entrez_fetch(db="nuccore", rettype = "fasta", id=messn[has.both])
write(messn.seq, file="ncbi-messngr-wd40-fbox.fna")
Find the genes that encode the proteins
genes <- entrez_link(dbfrom = "protein", id=proteins[has.both], by_id=TRUE,
linkname="protein_gene")
ugenes <- sapply(genes, function(m) m$links$protein_gene)
Find expression data for those genes
geoprof <- entrez_link(dbfrom = "gene", id=ugenes, by_id=TRUE,
linkname="gene_geoprofiles")
profiles <- lapply(geoprof, function(m) m$links$gene_geoprofiles)
ugeoprof <- unique(unlist(profiles))
geoprofiles_gds <- entrez_link(dbfrom = "geoprofiles", id=ugeoprof,
linkname="geoprofiles_gds")
Find papers associates with these genes
gene_pubmed <- entrez_link(dbfrom = "gene", id=ugenes, by_id=TRUE,
linkname="gene_pubmed")
upubmed <- unique(unlist(lapply(gene_pubmed, function(m) m$links$gene_pubmed)))
recs <- entrez_fetch(db="pubmed", id=upubmed, rettype="xml")
papers <- parse_pubmed_xml(recs)
Final result
sonuc <- data.frame(proteins,
name=pnames[proteins],
messn=sapply(messn, function(m) ifelse(is.null(m),"",m[1])),
domains=prot.domains,
has.fbox, has.wd40, has.both,
stringsAsFactors = FALSE)