April 19th, 2016
Welcome
to “Computing for Molecular Biology 2”
Plan for Today
- Midterm exam review
- Sequence similarity
- Sequence assembly
Midterm exam review
Please answer at least 3 of the following questions:
- What is Markdown? Why it is useful for Science?
- What are the elements of a structured document?
- What are the elements of this document? (the exam itself)
- Why it is important to separate content from style in a scientific report?
Please answer the following questions with your own words:
- What is GEO? Why is it useful for Molecular Biology and Genetics?
- What is MIAME?
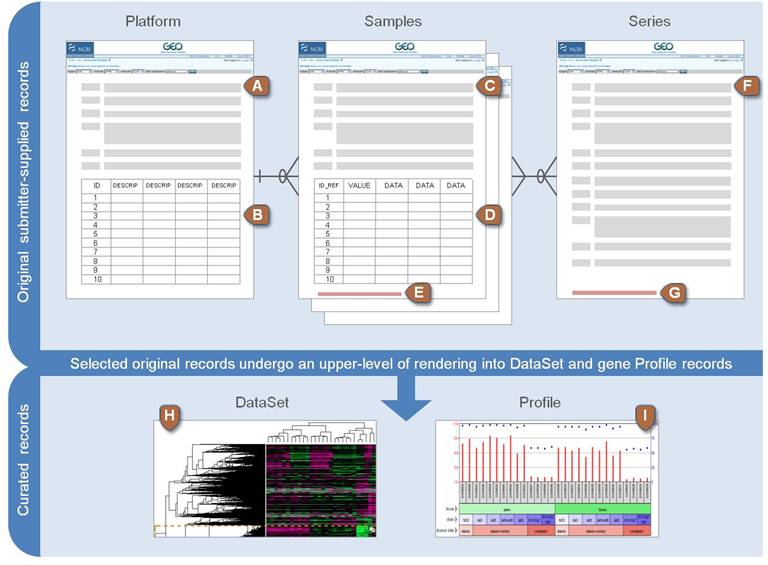
Explain the Entity-Relationship of GEO
Please describe it with your own words, considering:
- Which are all the entities?
- What are their relationships? That is, how are they related?
- What are their attributes? Remember that entities and relationships can have attributes
- What are the identifiers?
- What are the cardinalities?
Entity-Relationship Diagram of GEO
Please answer the following questions with your own words:
- Why clustering is an useful tool for Molecular Biology and Genetics?
- What is a distance in hierarchical clustering? Please give examples
- What is linkage? Please give examples
Please describe which file format is better suited for storing the following data and why:
- The sequences of amino-acids of a set of proteins
- The genome of a yeast and the location of its genes
- The locations in the chromosome of transcription factors binding sites
- The sequence of a plasmid
With your own words, please describe:
- How do you determine the origin of replication in a bacterial chromosome? Can you do the same in Eukarya? Why?
- How do you determine the binding sites of a transcription factor?
- What is a Motif? What is a Position Specific Score Matrix (also known as Position Weight Matrix)?
Please write an R function to transform a list of CDS (nucleotides) into a list of the aminoacidic sequences of the proteins they encode.
- Input: list of vector of characters named
CDS - Output: list of vector of characters
- you can use the function
translate()that transform a single CDS into the corresponding protein
Please write an R function to evaluate the score of each position of the genome given a PSSM
- Write first a function
score.pos()to evaluate the score of a fixed position. The inputs arepos: position in the genomegenome: vector of charsmat: a position specific score matrix
- Then write the code to evaluate
score.pos()on each position of a genome. The final output is a vector of numbers, each one representing the score of each position.
Sequence similarity
What is the function of this gene?
Sequence evolution
DNA replication is not perfect. Some bases can change
- Substitution of nucleotides
- Insertion
- Deletion
If these changes are lethal, the cell dies (by definition)
Therefore we only see changes that are compatible with cell life
This is one component of natural selection
Sequence and Function
Naturally, if two genes have the same sequence, they encode the same protein
If they differ in a few bases, the proteins will also differ a little, or less (why?)
So if two proteins are very similar, they probably do the same function
A few changes will probably not change they way it folds
Same shape, same function.
Comparing sequences