March 22th, 2016
Welcome
to “Computing for Molecular Biology 2”
Plan for Today
- What is an ORF? What is a CDS?
- How can we read a GFF file on R?
- How can we combine a FASTA file and a GFF file to get the gene sequences?
- What is a Transcription Factor (TF)?
- What is a Binding Site (BS)?
- What is a Motif? What is a Regular Expression?
- What is a Position Specific Score Matrix?
- How do we find Transcription Factor Binding Sites?
What is an ORF? What is a CDS?
What is an ORF? What is a CDS?
- Reading Frame: 6 ways of translate DNA to AA
- Open Reading Frame:
Start-Not Stop{repeated}-Stop - CDS: ORF that is translated
\[\{x\text{ is CDS}\}\subset\{x\text{ is ORF}\}\]
How can you find all ORFs of E.coli?
What are the inputs?
What is the output?
What are the steps between them?
Which ones are CDS?
CDS can be read from a GFF file
How can we read a GFF file on R?
gff <- read.delim("NC_000913.gff", header=F, comment.char="#")
summary(gff)
V1 V2 V3 V4
NC_000913.3:9720 RefSeq:9720 gene :4516 Min. : 1
CDS :4382 1st Qu.:1162247
repeat_region: 355 Median :2298492
exon : 178 Mean :2313290
tRNA : 89 3rd Qu.:3453929
ncRNA : 65 Max. :4640942
(Other) : 135
V5 V6 V7 V8
Min. : 255 .:9720 -:4682 .:5338
1st Qu.:1163115 +:5038 0:4370
Median :2299578 1: 7
Mean :2314649 2: 5
3rd Qu.:3455647
Max. :4641652
V9
ID=cds1219;Parent=gene1262;Note=pseudogene%2C transposase homolog;Dbxref=ASAP:ABE-0004159,ASAP:ABE-0004161,ASAP:ABE-0285106,UniProtKB%2FSwiss-Prot:P30192,EcoGene:EG11611,GeneID:4056037;gbkey=CDS;gene=insZ;pseudo=true;transl_table=11 : 3
ID=cds1389;Parent=gene1435;Note=pseudogene%2C autotransporter homolog%7Einterrupted by IS2 and IS30;Dbxref=ASAP:ABE-0004680,ASAP:ABE-0004694,ASAP:ABE-0285093,UniProtKB%2FSwiss-Prot:P33666,EcoGene:EG11307,GeneID:2847750;gbkey=CDS;gene=ydbA;pseudo=true;transl_table=11: 3
ID=cds1922;Parent=gene1981;Note=pseudogene%2C IpaH%2FYopM family;Dbxref=ASAP:ABE-0006435,ASAP:ABE-0006437,ASAP:ABE-0006440,ASAP:ABE-0285096,UniProtKB%2FSwiss-Prot:P76321,EcoGene:EG13281,GeneID:2847704;gbkey=CDS;gene=yedN;pseudo=true;transl_table=11 : 3
ID=cds3953;Parent=gene4120;Note=pseudogene%2C SopA-related%2C pentapeptide repeats-containing;Dbxref=ASAP:ABE-0013224,UniProtKB%2FSwiss-Prot:P32690,EcoGene:EG11927,GeneID:948546;gbkey=CDS;gene=yjbI;pseudo=true;transl_table=11 : 3
ID=cds1153;Parent=gene1190;Note=pseudogene;Dbxref=ASAP:ABE-0003933,ASAP:ABE-0285042,UniProtKB%2FSwiss-Prot:P76000,EcoGene:EG13890,GeneID:1450255;gbkey=CDS;gene=ycgI;pseudo=true;transl_table=11 : 2
ID=cds1302;Parent=gene1345;Note=pseudogene%7Eputative ATP-binding component of a transport system;Dbxref=ASAP:ABE-0004422,ASAP:ABE-0285045,UniProtKB%2FSwiss-Prot:P77481,EcoGene:EG13919,GeneID:4306141;gbkey=CDS;gene=ycjV;pseudo=true;transl_table=11 : 2
(Other) :9704
Output in FASTA format
How can we combine a FASTA file and a GFF file to get the gene sequences?
Write a function that combines a FASTA of the full genome and a GFF.
The output must be a FASTA file with the aminoacidic sequence of the coded proteins.
- What are the inputs?
- What is the output?
- What are the steps between them?
See write.fasta()
Transcription Regulation
Turning genes on and off
- What is a Transcription Factor (TF)?
- What is a Binding Site (BS)?
Regulation Mechanism
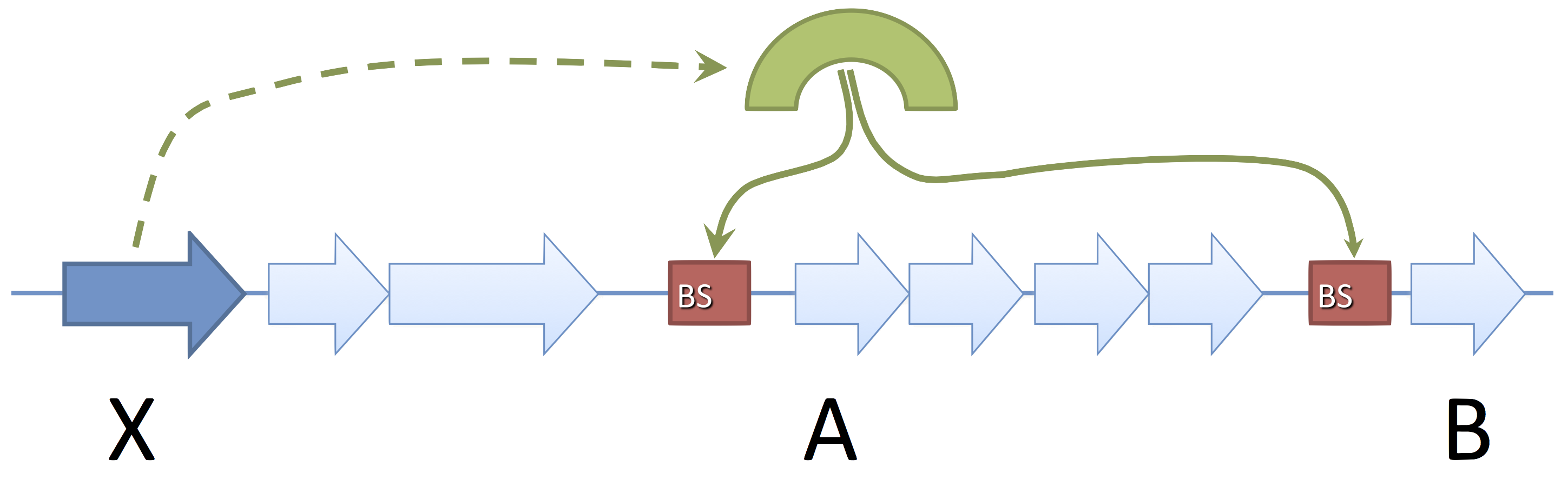
- Gene X codes for a Transcription Factor (TF)
- The TF attaches to DNA on binding sites (BS)
- This modifies the expression of genes A and B
- We say that X regulates A and B
What is a Motif? What is a Regular Expression?
Sigma 70 factor of E.coli binds to:
TTGACA-N(15-19)-TATAAT
What does this mean?
RegulonDB
The most complete database of E.coli regulation
Look for Sigma Factor 70
See also: Downloads
One TF, many BS
Transcription Factor Dan has 5 binding sites in different parts of the genome.
The sequences are:
GTTAATT GTGTATT ATTCATT GTTGATT GTTAATT
How do we summrize this?
How to make an “average”? A model?
Regular expression
A string to represent many strings
GTTAATT GTGTATT ATTCATT [GA]T[TG][ACTG]ATT GTTGATT GTTAATT
Position Specific Score Matrix
GTTAATT GTGTATT ATTCATT GTTGATT GTTAATT A | 1 0 0 2 5 0 0 C | 0 0 0 1 0 0 0 G | 4 0 1 1 0 0 0 T | 0 5 4 1 0 5 5
Each column has different score. Total score is the sum of all
How do we find Transcription Factor Binding Sites?
PSSM gives the Score of each position in the window
M[nucl,pos]
For each start position we evaluate the sum of the scores of the nucleotides in the window
Homework
Write a function to evaluate the score of a position for a given matrix
Inputs:
pos: position in the genomegenome: vector of charsmat: a position specific score matrix
Output: the score
Evaluate it on each position of E.coli genome.