March 15th, 2016
Welcome
to “Computing for Molecular Biology 2”
Plan for Today
- What is the genetic code?
- How was it discovered?
- Why George Gamow was wrong?
- Given the FASTA of a prokaryotic coding gene, How can you get the sequence of the protein?
- Write a function to transform the sequence of a gene into the corresponding protein
- How can we combine a FASTA file and a GFF file to get the gene sequences?
- How can we read a GFF file on R?
- What is an ORF? What is a CDS?
What is the genetic code?
How was it discovered?
How many genetic codes exist?
Why George Gamow was wrong?
library(seqinr) tablecode() SEQINR.UTIL$CODON.AA
Standard Genetic Code
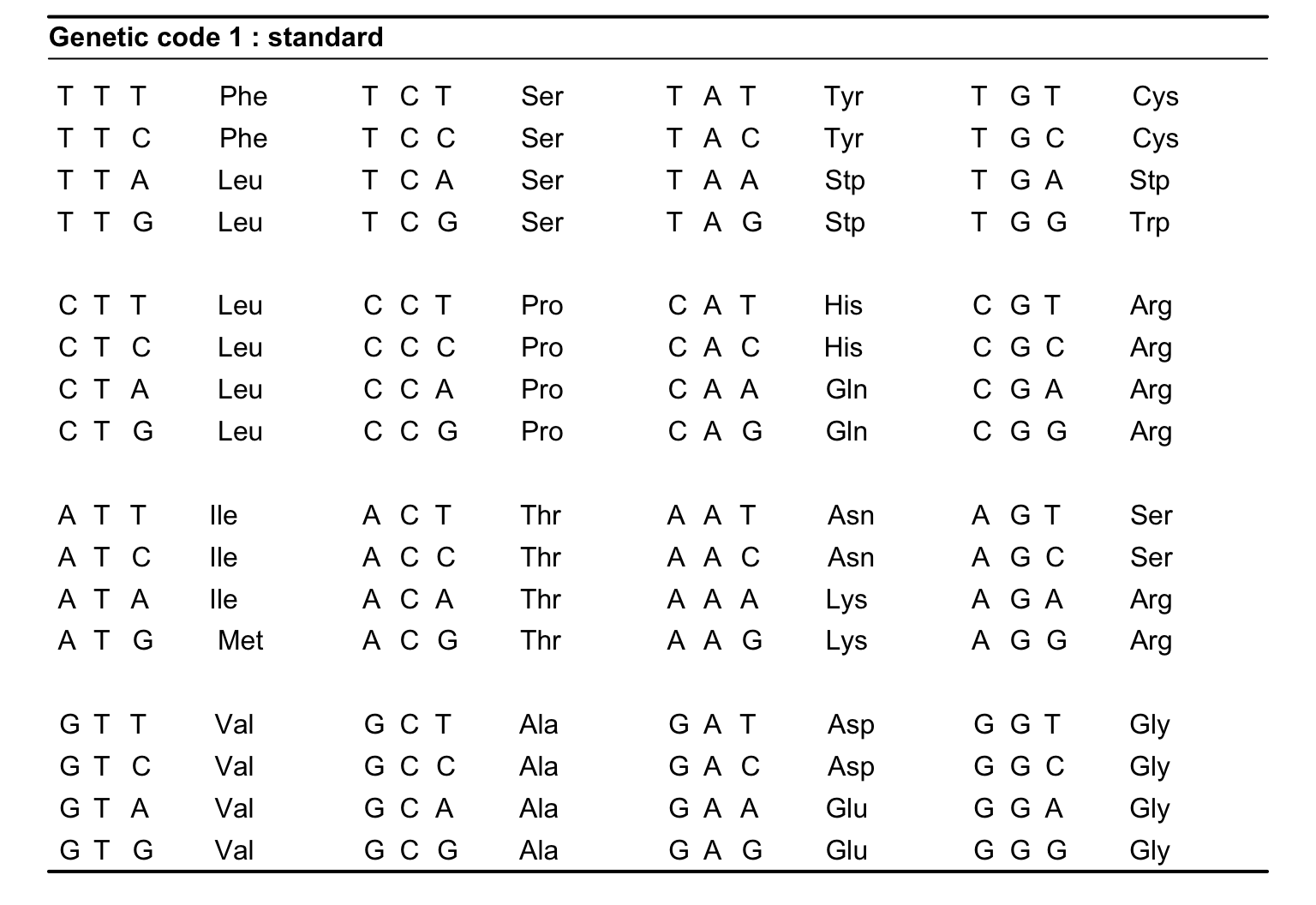
How can you get the sequence of the protein?
Given the FASTA of a prokaryotic coding gene?
- Using a function
- What are the inputs?
- What is the output?
- What are the steps between them?
Go ahead and write it
- Go to
http://www.ncbi.nlm.nih.gov/nuccore/NC_000913.3 - Choose Send -> Coding Sequences -> FASTA Nucleotide
- Rename the file as
NC_000913.ffn. Put it in a good folder. - You may also download the complete genome in a separate file
- Write the function in R
- The function
splitseqmay be useful
Homework
Codon Adaptation Index
(deadline: midterm)
- Write a function that takes a list of sequences of genes and counts the number of times each codon is used
- What should be the output of this function?
- Write the definition of CAI
- What is it useful for?
- Describe how to calculate it for each gene
- Write an R function that takes a list of sequences of genes and calculates CAI for each of them.
- Write an RMarkdown document to report the genes with atypical CAI on E.coli
Hint: The function syncodons() may be useful
Next week
Transcriptional Regulation
- What is a Transcription Factor (TF)?
- What is a Binding Site (BS)?
- What is a Motif? What is a Regular Expression?
- What is a Position Specific Score Matrix?
- How do we find Transcription Factor Binding Sites?
- Experimentally
- On the computer