March 1st, 2016
Clustering
Understanding by Forgetting
Identity
We recognize patterns, such as
- the same object from different angles

- the same object in different times


- objects of the same class


- hierarchy of classes


- abstractions of language: letters

- abstractions of sets: numbers
- abstractions of rules: algebra
Clustering
- split all the samples into meaningful classes
- Find the characteristic of each class
- classify all instances into classes
- determine the class of new instances
- determine the number of classes
A Correct Useful Clustering
- Different groupings can be correct at the same time
- The number of clusters depending on the context
- This is called granularity level
- meaning “the size of the grains”
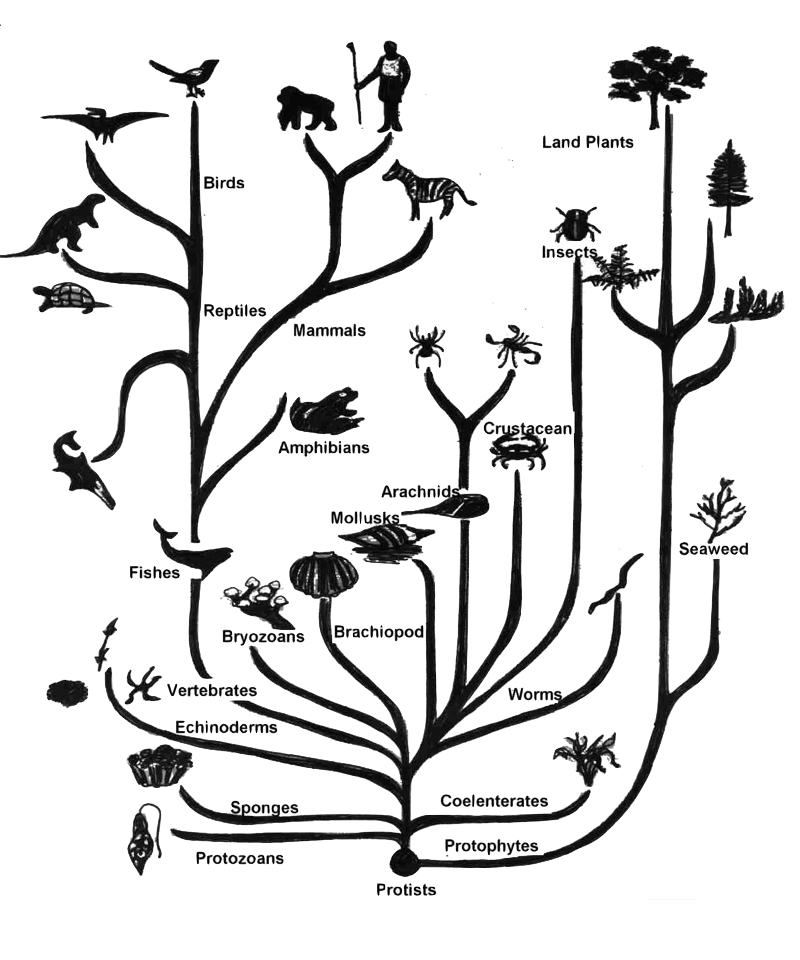
Tree of Life
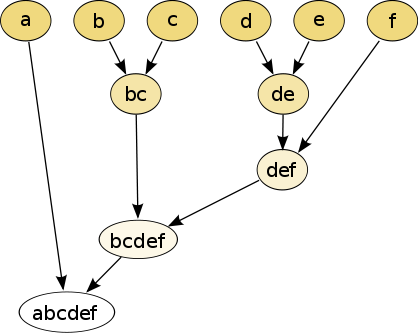
Hierarchical Clustering
Bottom up: joining one by one
- if \(\textrm{dist}(x, y)\) is the smallest distance, we join \(x\) and \(y\)
- we create cluster \(C\)
- we repeat until we have only one cluster
Application
microarray <- read.table("microarray.txt")
d <- dist(microarray, method="euclidean")
tree <- hclust(d, method = "average")
Euclidean Distance
\[\text{dist}_2(x,y)=\sqrt{(x_1-y_1)^2+\cdots +(x_n-y_n)^2}\]
Average Linkage
\[\text{dist}(x, C)=\text{mean} (\text{dist}(x, y): y \in C)\]
Hierarchical clustering
plot(tree, labels = FALSE)
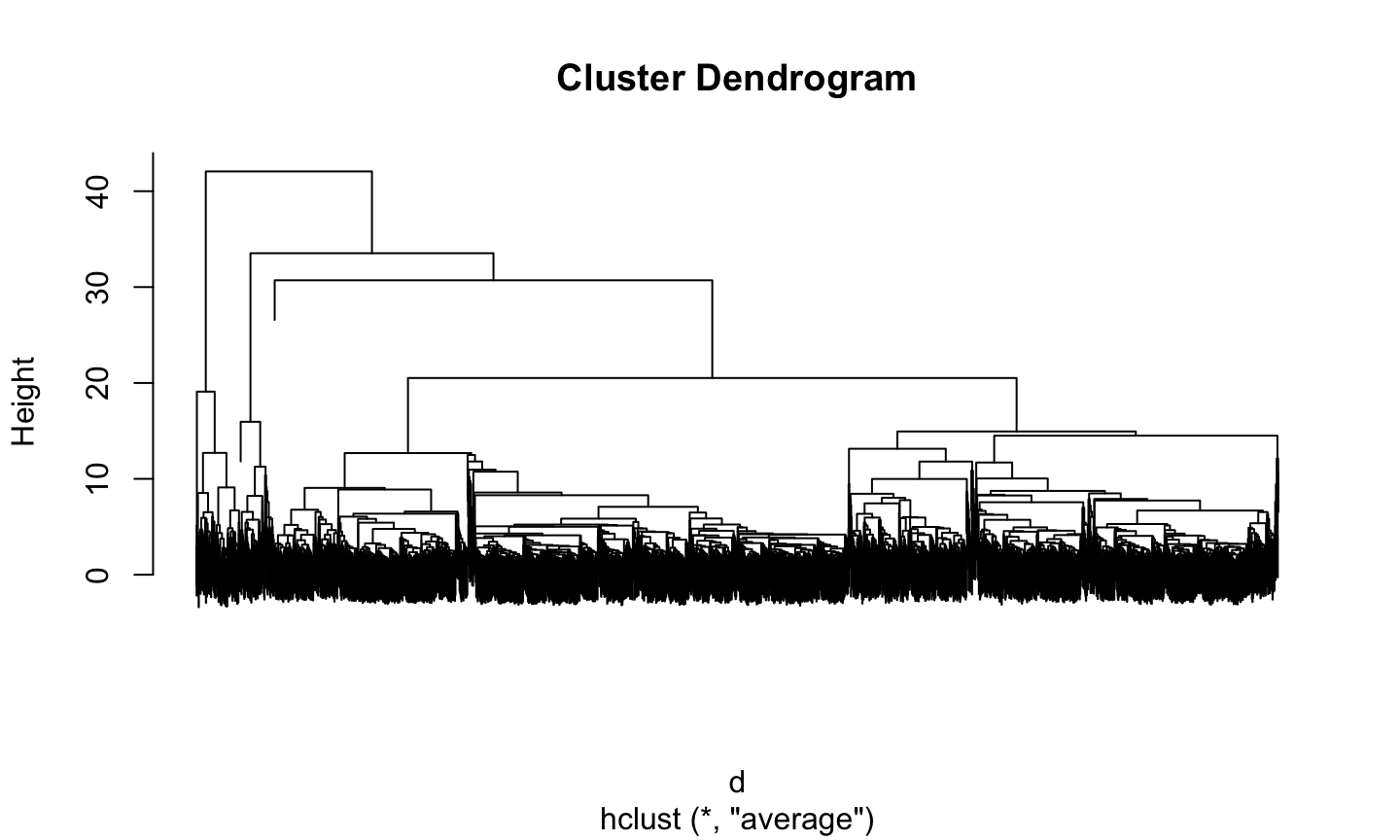
Which are the clusters?
Since this is a hierarchical clustering, the number of clusters depends on the distance threshold.
Higher cut value means less clusters
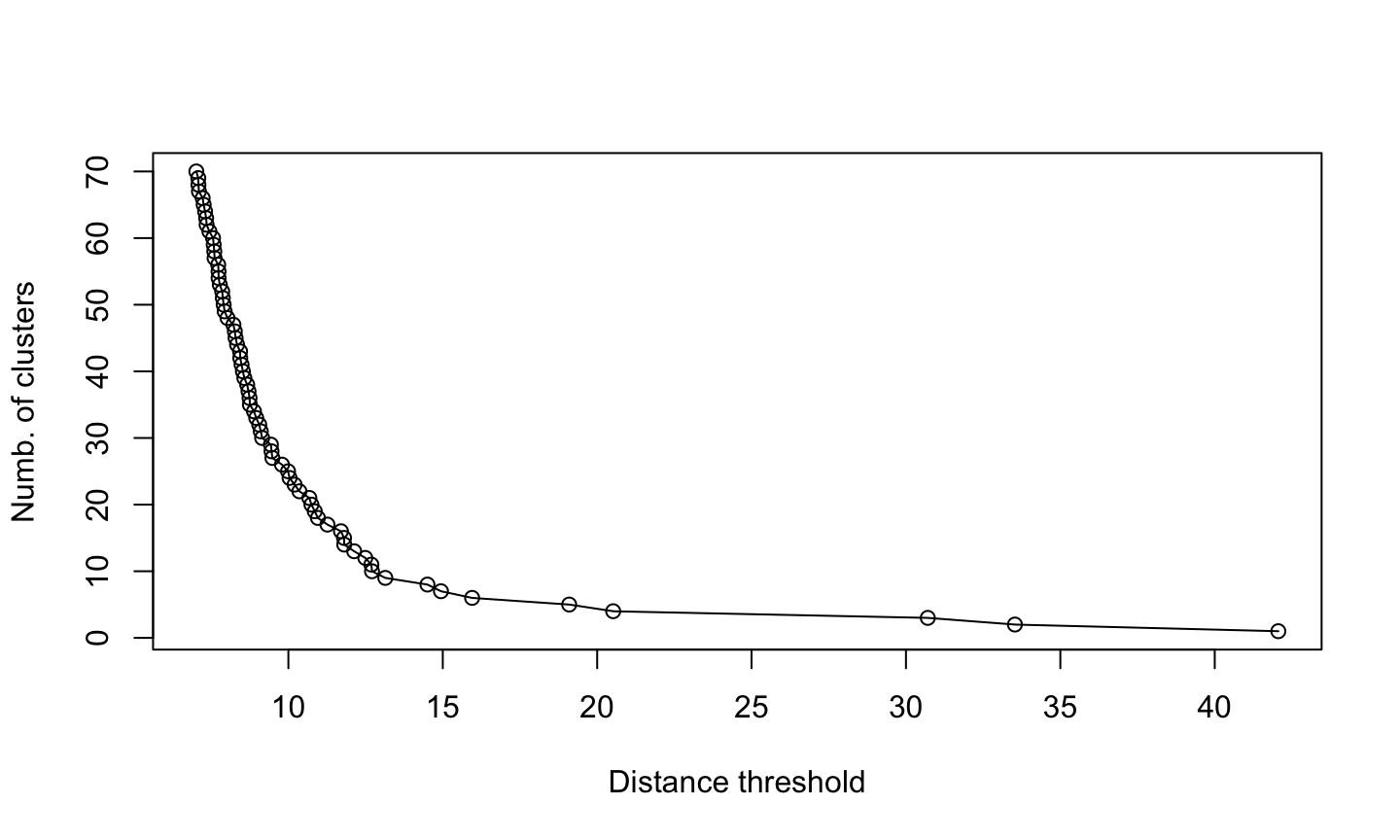
Less clusters means bigger clusters
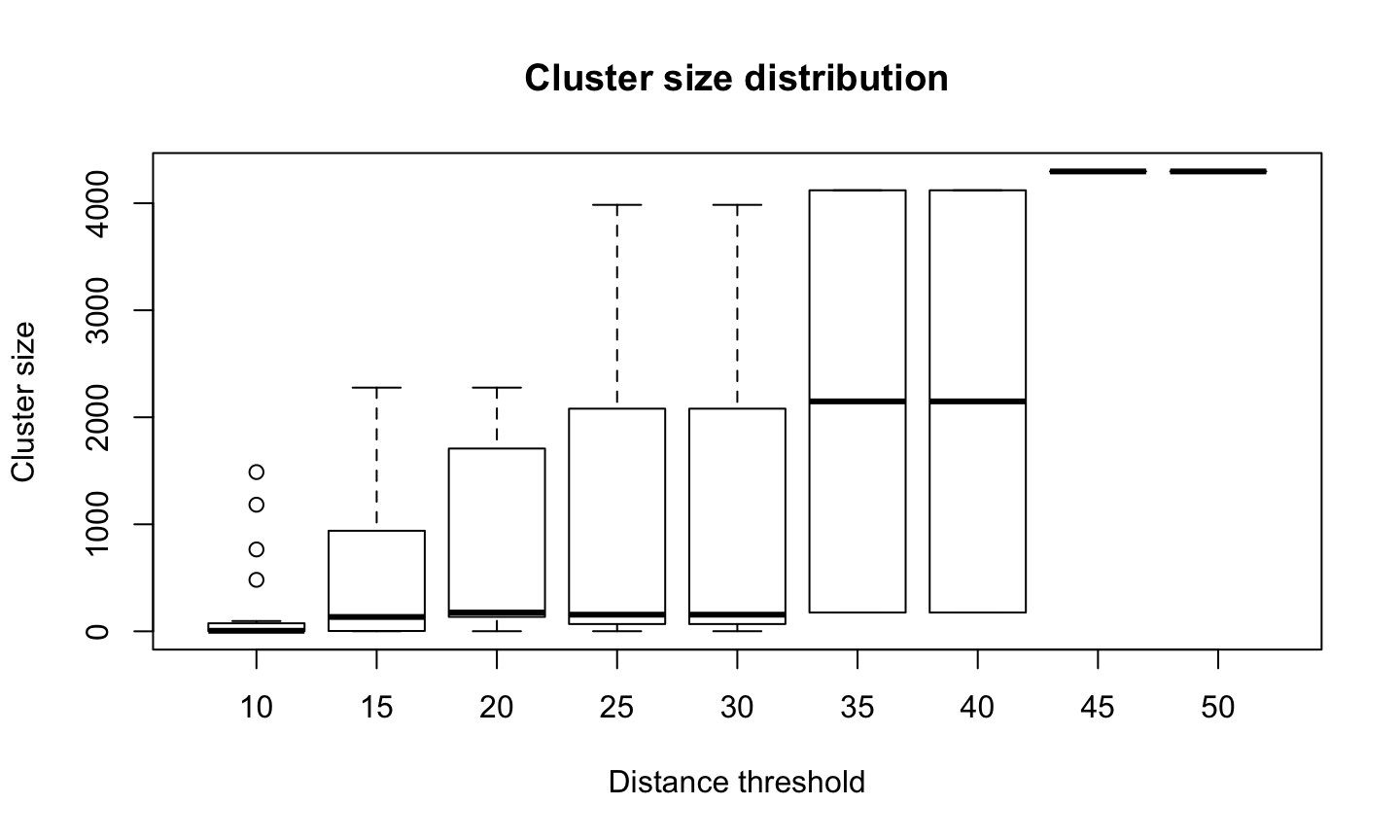
Doing the job
To finally split the samples into the clusters defined by the tree, we use the function cutree
cutree(tree, k = NULL, h = NULL)
- tree: a tree as produced by
hclust. - k: an integer scalar or vector with the desired number of groups
- h: numeric scalar or vector with heights where the tree should be cut
- sounds like cut tree
Example
cluster <- cutree(tree, h = 5) cluster
b4634 b3241 b3240 b3243 b3242 b2836 b0885 b1338 b1337 b1339
1 2 3 4 3 1 5 6 3 3
[ reached getOption("max.print") -- omitted 4287 entries ]
table(cluster)
cluster
1 2 3 4 5 6 7 8 9 10
277 74 386 440 327 486 78 223 22 38
[ reached getOption("max.print") -- omitted 166 entries ]
Some clusters
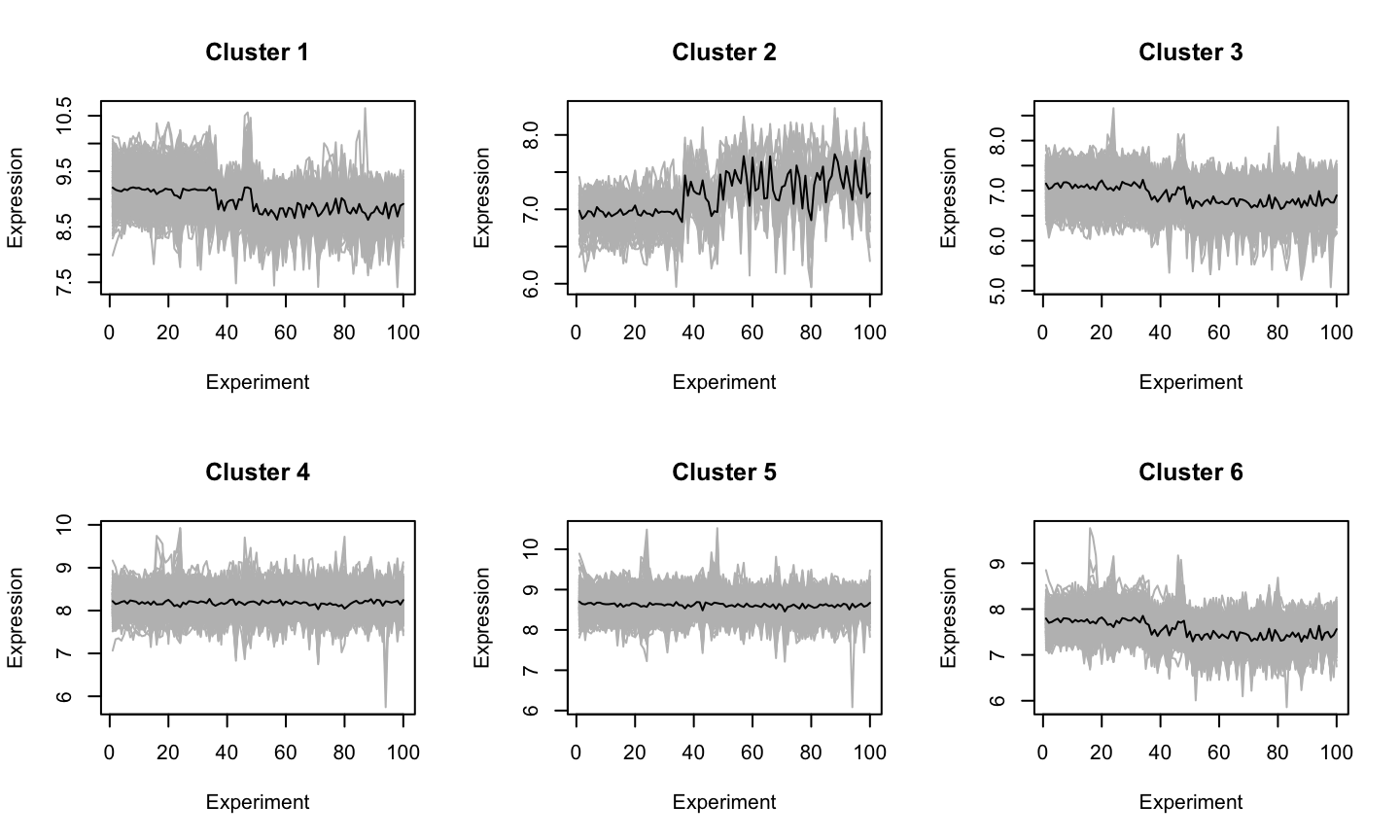
Genomic Sequences
FASTA file
>gi|556503834|ref|NC_000913.3| Escherichia coli str. K-12 substr. MG1655, complete genome AGCTTTTCATTCTGACTGCAACGGGCAATATGTCTCTGTGTGGATTAAAAAAAGAGTGTCTGATAGCAGC TTCTGAACTGGTTACCTGCCGTGAGTAAATTAAAATTTTATTGACTTAGGTCACTAAATACTTTAACCAA TATAGGCATAGCGCACAGACAGATAAAAATTACAGAGTACACAACATCCATGAAACGCATTAGCACCACC ATTACCACCACCATCACCATTACCACAGGTAACGGTGCGGGCTGACGCGTACAGGAAACACAGAAAAAAG CCCGCACCTGACAGTGCGGGCTTTTTTTTTCGACCAAAGGTAACGAGGTAACAACCATGCGAGTGTTGAA GTTCGGCGGTACATCAGTGGCAAATGCAGAACGTTTTCTGCGTGTTGCCGATATTCTGGAAAGCAATGCC AGGCAGGGGCAGGTGGCCACCGTCCTCTCTGCCCCCGCCAAAATCACCAACCACCTGGTGGCGATGATTG AAAAAACCATTAGCGGCCAGGATGCTTTACCCAATATCAGCGATGCCGAACGTATTTTTGCCGAACTTTT GACGGGACTCGCCGCCGCCCAGCCGGGGTTCCCGCTGGCGCAATTGAAAACTTTCGTCGATCAGGAATTT GCCCAAATAAAACATGTCCTGCATGGCATTAGTTTGTTGGGGCAGTGCCCGGATAGCATCAACGCTGCGC TGATTTGCCGTGGCGAGAAAATGTCGATCGCCATTATGGCCGGCGTATTAGAAGCGCGCGGTCACAACGT TACTGTTATCGATCCGGTCGAAAAACTGCTGGCAGTGGGGCATTACCTCGAATCTACCGTCGATATTGCT GAGTCCACCCGCCGTATTGCGGCAAGCCGCATTCCGGCTGATCACATGGTGCTGATGGCAGGTTTCACCG CCGGTAATGAAAAAGGCGAACTGGTGGTGCTTGGACGCAACGGTTCCGACTACTCTGCTGCGGTGCTGGC TGCCTGTTTACGCGCCGATTGTTGCGAGATTTGGACGGACGTTGACGGGGTCTATACCTGCGACCCGCGT CAGGTGCCCGATGCGAGGTTGTTGAAGTCGATGTCCTACCAGGAAGCGATGGAGCTTTCCTACTTCGGCG CTAAAGTTCTTCACCCCCGCACCATTACCCCCATCGCCCAGTTCCAGATCCCTTGCCTGATTAAAAATAC CGGAAATCCTCAAGCACCAGGTACGCTCATTGGTGCCAGCCGTGATGAAGACGAATTACCGGTCAAGGGC ATTTCCAATCTGAATAACATGGCAATGTTCAGCGTTTCTGGTCCGGGGATGAAAGGGATGGTCGGCATGG CGGCGCGCGTCTTTGCAGCGATGTCACGCGCCCGTATTTCCGTGGTGCTGATTACGCAATCATCTTCCGA ATACAGCATCAGTTTCTGCGTTCCACAAAGCGACTGTGTGCGAGCTGAACGGGCAATGCAGGAAGAGTTC TACCTGGAACTGAAAGAAGGCTTACTGGAGCCGCTGGCAGTGACGGAACGGCTGGCCATTATCTCGGTGG TAGGTGATGGTATGCGCACCTTGCGTGGGATCTCGGCGAAATTCTTTGCCGCACTGGCCCGCGCCAATAT CAACATTGTCGCCATTGCTCAGGGATCTTCTGAACGCTCAATCTCTGTCGTGGTAAATAACGATGATGCG ACCACTGGCGTGCGCGTTACTCATCAGATGCTGTTCAATACCGATCAGGTTATCGAAGTGTTTGTGATTG GCGTCGGTGGCGTTGGCGGTGCGCTGCTGGAGCAACTGAAGCGTCAGCAAAGCTGGCTGAAGAATAAACA TATCGACTTACGTGTCTGCGGTGTTGCCAACTCGAAGGCTCTGCTCACCAATGTACATGGCCTTAATCTG GAAAACTGGCAGGAAGAACTGGCGCAAGCCAAAGAGCCGTTTAATCTCGGGCGCTTAATTCGCCTCGTGA AAGAATATCATCTGCTGAACCCGGTCATTGTTGACTGCACTTCCAGCCAGGCAGTGGCGGATCAATATGC CGACTTCCTGCGCGAAGGTTTCCACGTTGTCACGCCGAACAAAAAGGCCAACACCTCGTCGATGGATTAC TACCATCAGTTGCGTTATGCGGCGGAAAAATCGCGGCGTAAATTCCTCTATGACACCAACGTTGGGGCTG GATTACCGGTTATTGAGAACCTGCAAAATCTGCTCAATGCAGGTGATGAATTGATGAAGTTCTCCGGCAT TCTTTCTGGTTCGCTTTCTTATATCTTCGGCAAGTTAGACGAAGGCATGAGTTTCTCCGAGGCGACCACG CTGGCGCGGGAAATGGGTTATACCGAACCGGACCCGCGAGATGATCTTTCTGGTATGGATGTGGCGCGTA AACTATTGATTCTCGCTCGTGAAACGGGACGTGAACTGGAGCTGGCGGATATTGAAATTGAACCTGTGCT GCCCGCAGAGTTTAACGCCGAGGGTGATGTTGCCGCTTTTATGGCGAATCTGTCACAACTCGACGATCTC TTTGCCGCGCGCGTGGCGAAGGCCCGTGATGAAGGAAAAGTTTTGCGCTATGTTGGCAATATTGATGAAG ATGGCGTCTGCCGCGTGAAGATTGCCGAAGTGGATGGTAATGATCCGCTGTTCAAAGTGAAAAATGGCGA AAACGCCCTGGCCTTCTATAGCCACTATTATCAGCCGCTGCCGTTGGTACTGCGCGGATATGGTGCGGGC AATGACGTTACAGCTGCCGGTGTCTTTGCTGATCTGCTACGTACCCTCTCATGGAAGTTAGGAGTCTGAC ATGGTTAAAGTTTATGCCCCGGCTTCCAGTGCCAATATGAGCGTCGGGTTTGATGTGCTCGGGGCGGCGG TGACACCTGTTGATGGTGCATTGCTCGGAGATGTAGTCACGGTTGAGGCGGCAGAGACATTCAGTCTCAA CAACCTCGGACGCTTTGCCGATAAGCTGCCGTCAGAACCACGGGAAAATATCGTTTATCAGTGCTGGGAG CGTTTTTGCCAGGAACTGGGTAAGCAAATTCCAGTGGCGATGACCCTGGAAAAGAATATGCCGATCGGTT CGGGCTTAGGCTCCAGTGCCTGTTCGGTGGTCGCGGCGCTGATGGCGATGAATGAACACTGCGGCAAGCC GCTTAATGACACTCGTTTGCTGGCTTTGATGGGCGAGCTGGAAGGCCGTATCTCCGGCAGCATTCATTAC GACAACGTGGCACCGTGTTTTCTCGGTGGTATGCAGTTGATGATCGAAGAAAACGACATCATCAGCCAGC AAGTGCCAGGGTTTGATGAGTGGCTGTGGGTGCTGGCGTATCCGGGGATTAAAGTCTCGACGGCAGAAGC CAGGGCTATTTTACCGGCGCAGTATCGCCGCCAGGATTGCATTGCGCACGGGCGACATCTGGCAGGCTTC ATTCACGCCTGCTATTCCCGTCAGCCTGAGCTTGCCGCGAAGCTGATGAAAGATGTTATCGCTGAACCCT ACCGTGAACGGTTACTGCCAGGCTTCCGGCAGGCGCGGCAGGCGGTCGCGGAAATCGGCGCGGTAGCGAG CGGTATCTCCGGCTCCGGCCCGACCTTGTTCGCTCTGTGTGACAAGCCGGAAACCGCCCAGCGCGTTGCC
FASTA file (amino acids)
>NC_000913.3_prot_NP_414542.1_1 [gene=thrL] [protein=thr operon leader peptide] [protein_id=NP_414542.1] [location=190..255] MKRISTTITTTITITTGNGAG >NC_000913.3_prot_NP_414543.1_2 [gene=thrA] [protein=Bifunctional aspartokinase/homoserine dehydrogenase 1] [protein_id=NP_414543.1] [location=337..2799] MRVLKFGGTSVANAERFLRVADILESNARQGQVATVLSAPAKITNHLVAMIEKTISGQDALPNISDAERI FAELLTGLAAAQPGFPLAQLKTFVDQEFAQIKHVLHGISLLGQCPDSINAALICRGEKMSIAIMAGVLEA RGHNVTVIDPVEKLLAVGHYLESTVDIAESTRRIAASRIPADHMVLMAGFTAGNEKGELVVLGRNGSDYS AAVLAACLRADCCEIWTDVDGVYTCDPRQVPDARLLKSMSYQEAMELSYFGAKVLHPRTITPIAQFQIPC LIKNTGNPQAPGTLIGASRDEDELPVKGISNLNNMAMFSVSGPGMKGMVGMAARVFAAMSRARISVVLIT QSSSEYSISFCVPQSDCVRAERAMQEEFYLELKEGLLEPLAVTERLAIISVVGDGMRTLRGISAKFFAAL ARANINIVAIAQGSSERSISVVVNNDDATTGVRVTHQMLFNTDQVIEVFVIGVGGVGGALLEQLKRQQSW LKNKHIDLRVCGVANSKALLTNVHGLNLENWQEELAQAKEPFNLGRLIRLVKEYHLLNPVIVDCTSSQAV ADQYADFLREGFHVVTPNKKANTSSMDYYHQLRYAAEKSRRKFLYDTNVGAGLPVIENLQNLLNAGDELM KFSGILSGSLSYIFGKLDEGMSFSEATTLAREMGYTEPDPRDDLSGMDVARKLLILARETGRELELADIE IEPVLPAEFNAEGDVAAFMANLSQLDDLFAARVAKARDEGKVLRYVGNIDEDGVCRVKIAEVDGNDPLFK VKNGENALAFYSHYYQPLPLVLRGYGAGNDVTAAGVFADLLRTLSWKLGV >NC_000913.3_prot_NP_414544.1_3 [gene=thrB] [protein=homoserine kinase] [protein_id=NP_414544.1] [location=2801..3733] MVKVYAPASSANMSVGFDVLGAAVTPVDGALLGDVVTVEAAETFSLNNLGRFADKLPSEPRENIVYQCWE RFCQELGKQIPVAMTLEKNMPIGSGLGSSACSVVAALMAMNEHCGKPLNDTRLLALMGELEGRISGSIHY DNVAPCFLGGMQLMIEENDIISQQVPGFDEWLWVLAYPGIKVSTAEARAILPAQYRRQDCIAHGRHLAGF IHACYSRQPELAAKLMKDVIAEPYRERLLPGFRQARQAVAEIGAVASGISGSGPTLFALCDKPETAQRVA DWLGKNYLQNQEGFVHICRLDTAGARVLEN >NC_000913.3_prot_NP_414545.1_4 [gene=thrC] [protein=L-threonine synthase] [protein_id=NP_414545.1] [location=3734..5020] MKLYNLKDHNEQVSFAQAVTQGLGKNQGLFFPHDLPEFSLTEIDEMLKLDFVTRSAKILSAFIGDEIPQE ILEERVRAAFAFPAPVANVESDVGCLELFHGPTLAFKDFGGRFMAQMLTHIAGDKPVTILTATSGDTGAA VAHAFYGLPNVKVVILYPRGKISPLQEKLFCTLGGNIETVAIDGDFDACQALVKQAFDDEELKVALGLNS ANSINISRLLAQICYYFEAVAQLPQETRNQLVVSVPSGNFGDLTAGLLAKSLGLPVKRFIAATNVNDTVP RFLHDGQWSPKATQATLSNAMDVSQPNNWPRVEELFRRKIWQLKELGYAAVDDETTQQTMRELKELGYTS EPHAAVAYRALRDQLNPGEYGLFLGTAHPAKFKESVEAILGETLDLPKELAERADLPLLSHNLPADFAAL RKLMMNHQ >NC_000913.3_prot_NP_414546.1_5 [gene=yaaX] [protein=DUF2502 family putative periplasmic protein] [protein_id=NP_414546.1] [location=5234..5530] MKKMQSIVLALSLVLVAPMAAQAAEITLVPSVKLQIGDRDNRGYYWDGGHWRDHGWWKQHYEWRGNRWHL HGPPPPPRHHKKAPHDHHGGHGPGKHHR >NC_000913.3_prot_NP_414547.1_6 [gene=yaaA] [protein=peroxide resistance protein, lowers intracellular iron] [protein_id=NP_414547.1] [location=complement(5683..6459)] MLILISPAKTLDYQSPLTTTRYTLPELLDNSQQLIHEARKLTPPQISTLMRISDKLAGINAARFHDWQPD FTPANARQAILAFKGDVYTGLQAETFSEDDFDFAQQHLRMLSGLYGVLRPLDLMQPYRLEMGIRLENARG
GFF file
##gff-version 3 #!gff-spec-version 1.20 #!processor NCBI annotwriter ##sequence-region NC_000913.3 1 4641652 ##species http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=511145 NC_000913.3 RefSeq region 1 4641652 . + . ID=id0;Name=ANONYMOUS;Dbxref=taxon:511145;Is_circular=true;gbkey=Src;genome=chromosome;mol_type=genomic DNA;strain=K-12;substrain=MG1655 NC_000913.3 RefSeq gene 190 255 . + . ID=gene0;Name=thrL;Dbxref=EcoGene:EG11277,GeneID:944742;gbkey=Gene;gene=thrL;gene_synonym=ECK0001,JW4367;locus_tag=b0001 NC_000913.3 RefSeq CDS 190 255 . + 0 ID=cds0;Name=NP_414542.1;Parent=gene0;Dbxref=ASAP:ABE-0000006,UniProtKB%2FSwiss-Prot:P0AD86,Genbank:NP_414542.1,EcoGene:EG11277,GeneID:944742;gbkey=CDS;gene=thrL;product=thr operon leader peptide;protein_id=NP_414542.1;transl_table=11 NC_000913.3 RefSeq gene 337 2799 . + . ID=gene1;Name=thrA;Dbxref=EcoGene:EG10998,GeneID:945803;gbkey=Gene;gene=thrA;gene_synonym=ECK0002,Hs,JW0001,thrA1,thrA2,thrD;locus_tag=b0002 NC_000913.3 RefSeq CDS 337 2799 . + 0 ID=cds1;Name=NP_414543.1;Parent=gene1;Note=bifunctional: aspartokinase I %28N-terminal%29%3B homoserine dehydrogenase I %28C-terminal%29;Dbxref=ASAP:ABE-0000008,UniProtKB%2FSwiss-Prot:P00561,Genbank:NP_414543.1,EcoGene:EG10998,GeneID:945803;gbkey=CDS;gene=thrA;product=Bifunctional aspartokinase%2Fhomoserine dehydrogenase 1;protein_id=NP_414543.1;transl_table=11 NC_000913.3 RefSeq gene 2801 3733 . + . ID=gene2;Name=thrB;Dbxref=EcoGene:EG10999,GeneID:947498;gbkey=Gene;gene=thrB;gene_synonym=ECK0003,JW0002;locus_tag=b0003 NC_000913.3 RefSeq CDS 2801 3733 . + 0 ID=cds2;Name=NP_414544.1;Parent=gene2;Dbxref=ASAP:ABE-0000010,UniProtKB%2FSwiss-Prot:P00547,Genbank:NP_414544.1,EcoGene:EG10999,GeneID:947498;gbkey=CDS;gene=thrB;product=homoserine kinase;protein_id=NP_414544.1;transl_table=11 NC_000913.3 RefSeq gene 3734 5020 . + . ID=gene3;Name=thrC;Dbxref=EcoGene:EG11000,GeneID:945198;gbkey=Gene;gene=thrC;gene_synonym=ECK0004,JW0003;locus_tag=b0004 NC_000913.3 RefSeq CDS 3734 5020 . + 0 ID=cds3;Name=NP_414545.1;Parent=gene3;Dbxref=ASAP:ABE-0000012,UniProtKB%2FSwiss-Prot:P00934,Genbank:NP_414545.1,EcoGene:EG11000,GeneID:945198;gbkey=CDS;gene=thrC;product=L-threonine synthase;protein_id=NP_414545.1;transl_table=11 NC_000913.3 RefSeq gene 5234 5530 . + . ID=gene4;Name=yaaX;Dbxref=EcoGene:EG14384,GeneID:944747;gbkey=Gene;gene=yaaX;gene_synonym=ECK0005,JW0004;locus_tag=b0005 NC_000913.3 RefSeq CDS 5234 5530 . + 0 ID=cds4;Name=NP_414546.1;Parent=gene4;Dbxref=ASAP:ABE-0000015,UniProtKB%2FSwiss-Prot:P75616,Genbank:NP_414546.1,EcoGene:EG14384,GeneID:944747;gbkey=CDS;gene=yaaX;product=DUF2502 family putative periplasmic protein;protein_id=NP_414546.1;transl_table=11 NC_000913.3 RefSeq repeat_region 5565 5669 . + . ID=id1;Note=RIP1 %28repetitive extragenic palindromic%29 element%3B contains 2 REP sequences and 1 IHF site;gbkey=repeat_region NC_000913.3 RefSeq gene 5683 6459 . - . ID=gene5;Name=yaaA;Dbxref=EcoGene:EG10011,GeneID:944749;gbkey=Gene;gene=yaaA;gene_synonym=ECK0006,JW0005;locus_tag=b0006 NC_000913.3 RefSeq CDS 5683 6459 . - 0 ID=cds5;Name=NP_414547.1;Parent=gene5;Dbxref=ASAP:ABE-0000018,UniProtKB%2FSwiss-Prot:P0A8I3,Genbank:NP_414547.1,EcoGene:EG10011,GeneID:944749;gbkey=CDS;gene=yaaA;product=peroxide resistance protein%2C lowers intracellular iron;protein_id=NP_414547.1;transl_table=11 NC_000913.3 RefSeq gene 6529 7959 . - . ID=gene6;Name=yaaJ;Dbxref=EcoGene:EG11555,GeneID:944745;gbkey=Gene;gene=yaaJ;gene_synonym=ECK0007,JW0006;locus_tag=b0007 NC_000913.3 RefSeq CDS 6529 7959 . - 0 ID=cds6;Name=NP_414548.1;Parent=gene6;Note=inner membrane transport protein;Dbxref=ASAP:ABE-0000020,UniProtKB%2FSwiss-Prot:P30143,Genbank:NP_414548.1,EcoGene:EG11555,GeneID:944745;gbkey=CDS;gene=yaaJ;product=putative transporter;protein_id=NP_414548.1;transl_table=11 NC_000913.3 RefSeq gene 8238 9191 . + . ID=gene7;Name=talB;Dbxref=EcoGene:EG11556,GeneID:944748;gbkey=Gene;gene=talB;gene_synonym=ECK0008,JW0007,yaaK;locus_tag=b0008 NC_000913.3 RefSeq CDS 8238 9191 . + 0 ID=cds7;Name=NP_414549.1;Parent=gene7;Dbxref=ASAP:ABE-0000027,UniProtKB%2FSwiss-Prot:P0A870,Genbank:NP_414549.1,EcoGene:EG11556,GeneID:944748;gbkey=CDS;gene=talB;product=transaldolase B;protein_id=NP_414549.1;transl_table=11 NC_000913.3 RefSeq gene 9306 9893 . + . ID=gene8;Name=mog;Dbxref=EcoGene:EG11511,GeneID:944760;gbkey=Gene;gene=mog;gene_synonym=bisD,chlG,ECK0009,JW0008,mogA,yaaG;locus_tag=b0009 NC_000913.3 RefSeq CDS 9306 9893 . + 0 ID=cds8;Name=NP_414550.1;Parent=gene8;Note=putative molybdochetalase in molybdopterine biosynthesis;Dbxref=ASAP:ABE-0000030,UniProtKB%2FSwiss-Prot:P0AF03,Genbank:NP_414550.1,EcoGene:EG11511,GeneID:944760;gbkey=CDS;gene=mog;product=molybdochelatase incorporating molybdenum into molybdopterin;protein_id=NP_414550.1;transl_table=11 NC_000913.3 RefSeq gene 9928 10494 . - . ID=gene9;Name=satP;Dbxref=EcoGene:EG11512,GeneID:944792;gbkey=Gene;gene=satP;gene_synonym=ECK0010,JW0009,yaaH;locus_tag=b0010 NC_000913.3 RefSeq CDS 9928 10494 . - 0 ID=cds9;Name=NP_414551.1;Parent=gene9;Dbxref=ASAP:ABE-0000032,UniProtKB%2FSwiss-Prot:P0AC98,Genbank:NP_414551.1,EcoGene:EG11512,GeneID:944792;gbkey=CDS;gene=satP;product=succinate-acetate transporter;protein_id=NP_414551.1;transl_table=11 NC_000913.3 RefSeq gene 10643 11356 . - . ID=gene10;Name=yaaW;Dbxref=EcoGene:EG14340,GeneID:944771;gbkey=Gene;gene=yaaW;gene_synonym=ECK0011,JW0010;locus_tag=b0011 NC_000913.3 RefSeq CDS 10643 11356 . - 0 ID=cds10;Name=NP_414552.1;Parent=gene10;Dbxref=ASAP:ABE-0000037,UniProtKB%2FSwiss-Prot:P75617,Genbank:NP_414552.1,EcoGene:EG14340,GeneID:944771;gbkey=CDS;gene=yaaW;product=UPF0174 family protein;protein_id=NP_414552.1;transl_table=11 NC_000913.3 RefSeq gene 11382 11786 . - . ID=gene11;Name=yaaI;Dbxref=EcoGene:EG11513,GeneID:944751;gbkey=Gene;gene=yaaI;gene_synonym=ECK0013,JW0012;locus_tag=b0013 NC_000913.3 RefSeq CDS 11382 11786 . - 0 ID=cds11;Name=NP_414554.1;Parent=gene11;Dbxref=ASAP:ABE-0000043,UniProtKB%2FSwiss-Prot:P28696,Genbank:NP_414554.1,EcoGene:EG11513,GeneID:944751;gbkey=CDS;gene=yaaI;product=UPF0412 family protein;protein_id=NP_414554.1;transl_table=11 NC_000913.3 RefSeq gene 12163 14079 . + . ID=gene12;Name=dnaK;Dbxref=EcoGene:EG10241,GeneID:944750;gbkey=Gene;gene=dnaK;gene_synonym=ECK0014,groPAB,groPC,groPF,grpC,grpF,JW0013,seg;locus_tag=b0014 NC_000913.3 RefSeq CDS 12163 14079 . + 0 ID=cds12;Name=NP_414555.1;Parent=gene12;Note=chaperone Hsp70%3B DNA biosynthesis%3B autoregulated heat shock proteins;Dbxref=ASAP:ABE-0000052,UniProtKB%2FSwiss-Prot:P0A6Y8,Genbank:NP_414555.1,EcoGene:EG10241,GeneID:944750;gbkey=CDS;gene=dnaK;product=chaperone Hsp70%2C with co-chaperone DnaJ;protein_id=NP_414555.1;transl_table=11 NC_000913.3 RefSeq gene 14168 15298 . + . ID=gene13;Name=dnaJ;Dbxref=EcoGene:EG10240,GeneID:944753;gbkey=Gene;gene=dnaJ;gene_synonym=ECK0015,faa,groP,grpC,JW0014;locus_tag=b0015 NC_000913.3 RefSeq CDS 14168 15298 . + 0 ID=cds13;Name=NP_414556.1;Parent=gene13;Note=chaperone with DnaK%3B heat shock protein;Dbxref=ASAP:ABE-0000054,UniProtKB%2FSwiss-Prot:P08622,Genbank:NP_414556.1,EcoGene:EG10240,GeneID:944753;gbkey=CDS;gene=dnaJ;product=chaperone Hsp40%2C DnaK co-chaperone;protein_id=NP_414556.1;transl_table=11
GenBank file (partial)
LOCUS NC_000913 4641652 bp DNA circular CON 15-MAY-2014
DEFINITION Escherichia coli str. K-12 substr. MG1655, complete genome.
ACCESSION NC_000913
VERSION NC_000913.3 GI:556503834
DBLINK BioProject: PRJNA57779
BioSample: SAMN02604091
KEYWORDS RefSeq.
SOURCE Escherichia coli str. K-12 substr. MG1655
ORGANISM Escherichia coli str. K-12 substr. MG1655
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales;
Enterobacteriaceae; Escherichia.
FEATURES Location/Qualifiers
source 1..4641652
/organism="Escherichia coli str. K-12 substr. MG1655"
/mol_type="genomic DNA"
/strain="K-12"
/sub_strain="MG1655"
/db_xref="taxon:511145"
gene 190..255
/gene="thrL"
/locus_tag="b0001"
/gene_synonym="ECK0001; JW4367"
/db_xref="EcoGene:EG11277"
/db_xref="GeneID:944742"
CDS 190..255
/gene="thrL"
/locus_tag="b0001"
/gene_synonym="ECK0001; JW4367"
ORIGIN
1 agcttttcat tctgactgca acgggcaata tgtctctgtg tggattaaaa aaagagtgtc
61 tgatagcagc ttctgaactg gttacctgcc gtgagtaaat taaaatttta ttgacttagg
121 tcactaaata ctttaaccaa tataggcata gcgcacagac agataaaaat tacagagtac
181 acaacatcca tgaaacgcat tagcaccacc attaccacca ccatcaccat taccacaggt
241 aacggtgcgg gctgacgcgt acaggaaaca cagaaaaaag cccgcacctg acagtgcggg
301 cttttttttt cgaccaaagg taacgaggta acaaccatgc gagtgttgaa gttcggcggt
361 acatcagtgg caaatgcaga acgttttctg cgtgttgccg atattctgga aagcaatgcc
421 aggcaggggc aggtggccac cgtcctctct gcccccgcca aaatcaccaa ccacctggtg
GC-content
From Wikipedia, the free encyclopedia
The percentage of nitrogenous bases on a DNA molecule that are either guanine or cytosine.
- GC content is found to be variable with different organisms.
- The committee on bacterial systematics has recommended use of GC ratios in higher level hierarchical classification
The GC-content can be measured by several means,
- Measuring the melting temperature of the DNA double helix using spectrophotometry
- The absorbance of DNA at a wavelength of 260 nm increases sharply when the double-stranded DNA separates into two single strands when sufficiently heated.
Determination of GC content
In alternative manner, if the DNA has been sequenced then the GC-content can be accurately calculated by simple arithmetic.
GC-content percentage is calculated as \[\frac{G+C}{A+T+G+C}\]
Exercise 1
Determine the GC content of E.coli
Is this GC content uniform through all genome?
Chargaff’s rules
From Wikipedia, the free encyclopedia
Discovered by Austrian chemist Erwin Chargaff in 1952.
DNA from any cell of all organisms has a 1:1 ratio of pyrimidine and purine bases
The amount of guanine is equal to cytosine and the amount of adenine is equal to thymine.
Elson D, Chargaff E (1952). On the deoxyribonucleic acid content of sea urchin gametes. Experientia 8 (4): 143–145
Chargaff E, Lipshitz R, Green C (1952). Composition of the deoxypentose nucleic acids of four genera of sea-urchin. J Biol Chem 195 (1): 155–160
Chargaff’s rules
First parity rule
A double-stranded DNA molecule globally has percentage base pair equality: \(\%A = \%T\) and \(\%G = \%C\)
- The rigorous validation of the rule constitutes the basis of Watson-Crick pairs
Second parity rule
Both \(\%A \approx \%T\) and \(\%G \approx \%C\) are valid for each of the two DNA strands
- This describes only a global feature of the base composition in a single DNA strand
Exercise 2
Why the first rule is always valid?
How do you determine if \(\%G \approx \%C\)?
Is this valid through all the genome?
GC Skew
What is the difference \(G-C\) respect to the total \(G+C\)?
Calculate the ratio \[\frac{G-C}{G+C}\] using sliding windows and plot it
What do you see?
Chargaff second rule
In each DNA strand the frequency of occurrence of G is equal to C because the substitution rate is presumably equal
Hence, the second parity rule only exists when there is no mutation or substitution
There is a richness of G over C and T over A in the leading strand
- and vice versa for the lagging strand
GC skew changes sign at the boundaries of the two replichores
This corresponds to DNA replication origin or terminus
Comparative Genometrics
Roten C-AH, Gamba P, Barblan J-L, Karamata D. Comparative Genometrics (CG): a database dedicated to biometric comparisons of whole genomes. Nucleic Acids Research. 2002;30(1):142-144
Credits of Images
Chair image by Alex Rio Brazil - Own work, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=8045709
dogs By
YellowLabradorLooking_new.jpg:derivative work: Djmirko (talk)YellowLabradorLooking.jpg:User:HabjGolden_Retriever_Sammy.jpg:Pharaoh HoundCockerpoo.jpg:ALMMLonghaired_yorkie.jpg:Ed Garcia fromUnited StatesBoxer_female_brown.jpg:Flickr userboxercabMilù_050.JPG:AleRBeagle1.jpg:TobycatBasset_Hound_600.jpg:ToBNewfoundland_dog_Smoky.jpg:Flickr user DanDee Shotsderivative work: December21st2012Freak (talk) -YellowLabradorLooking_new.jpgGolden_Retriever_Sammy.jpgCockerpoo.jpgLonghaired_yorkie.jpgBoxer_female_brown.jpgMilù_050.JPGBeagle1.jpgBasset_Hound_600.jpgNewfoundland_dog_Smoky.jpg, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=10793219Allegory of the cave By Veldkamp, Gabriele and Maurer, Markus - Veldkamp, Gabriele. Zukunftsorientierte Gestaltung informationstechnologischer Netzwerke im Hinblick auf die Handlungsfähigkeit des Menschen. Aachener Reihe Mensch und Technik, Band 15, Verlag der Augustinus Buchhandlung, Aachen 1996, Germany, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=24826744