May 10th, 2016
Welcome to the last class
of “Computing for Molecular Biology 2”
Plan for Today
- Experimental Design
- Simulation
Genome Assembly
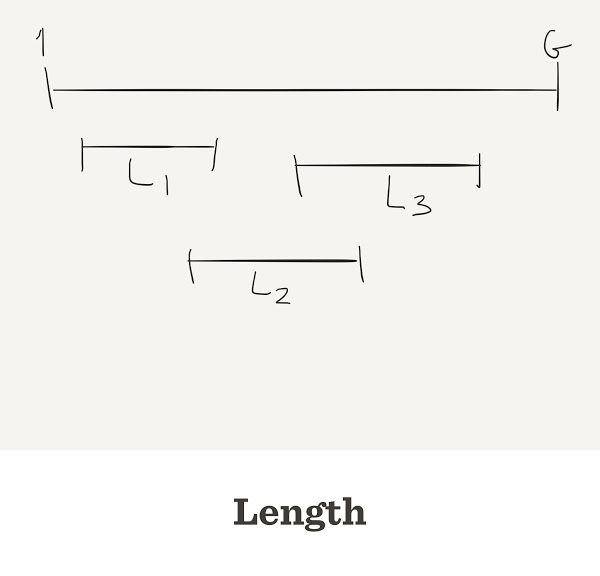
Genome Assembly
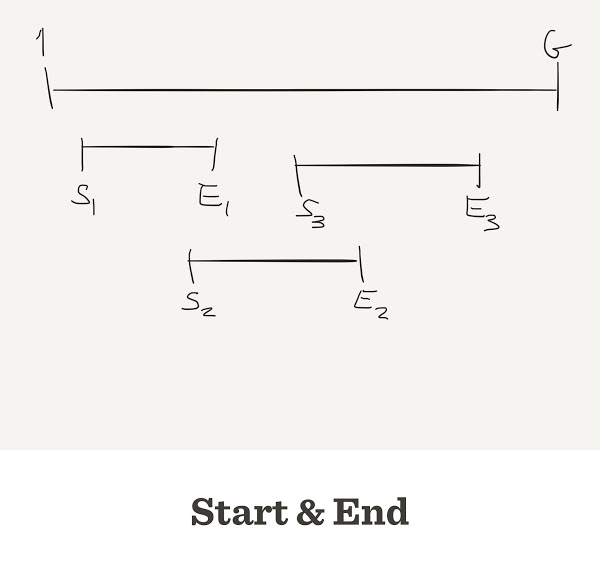
Genome Assembly
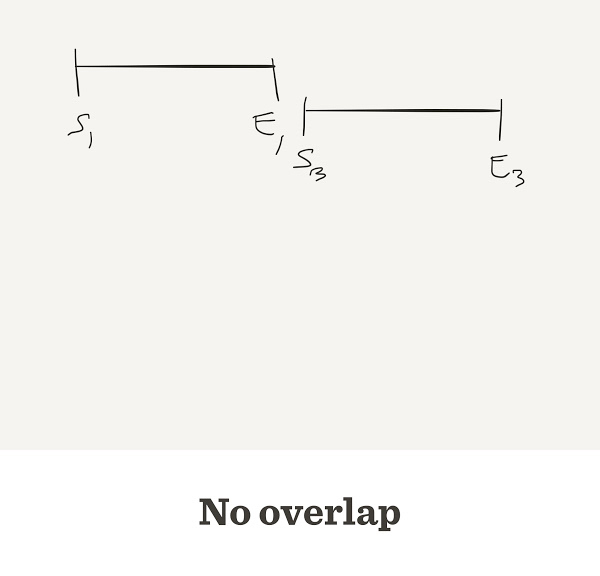
Genome Assembly
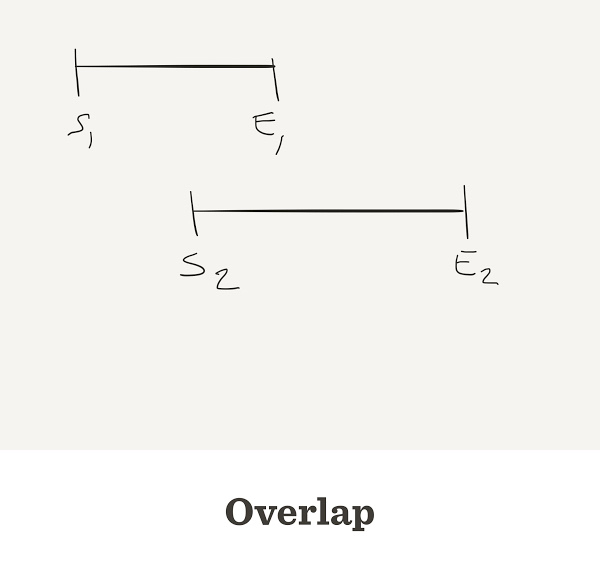
Genome Assembly
Gap counter
num.gaps <- function(G,L,n) {
start <- sample(G, n, replace=TRUE)
start <- sort(start)
end <- start + L
return(sum(start[-1] > end[-length(end)]))
}
- G: Length of genome
- L: Length of fragment/read
- n: number of fragments/reads
Gap counter
num.gaps <- function(G,L,n) {
start <- sample(G, n, replace=TRUE)
start <- sort(start)
end <- start + L
gap <- start[-1] - end[-length(end)]
return(sum(gap > 0))
}
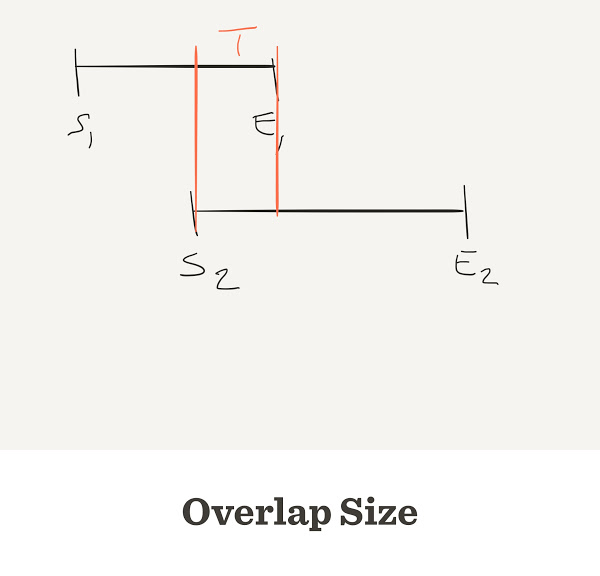
Gap counter
num.gaps <- function(G,L,n) {
start <- sample(G, n, replace=TRUE)
start <- sort(start)
end <- start + L
overlap <- end[-length(end)] - start[-1]
return(sum(overlap < 0))
}
Contig counter
num.gaps <- function(G,L,n,t) {
start <- sample(G, n, replace=TRUE)
start <- sort(start)
end <- start + L
overlap <- end[-length(end)] - start[-1]
return(sum(overlap < t))
}
- G: Length of genome
- L: Length of fragment/read
- n: number of fragments/reads
- t: minimum overlap length