Türkçe bilmiyorum 😟
I am
- New Assistant Professor at Molecular Biology and Genomics Department
- Mathematical Engineer, U. of Chile
- PhD Informatics, U Rennes 1, France
- PhD Mathematical Modeling, U. of Chile
- not a Biologist
- but an Applied Mathematician who can speak “biologist language”

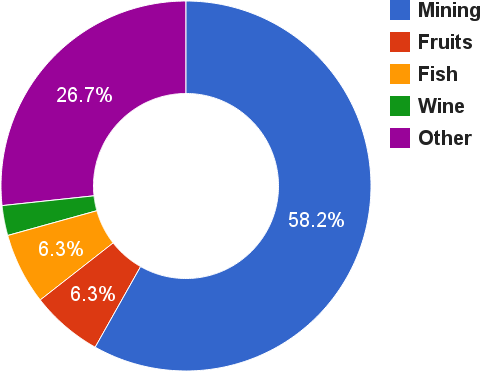

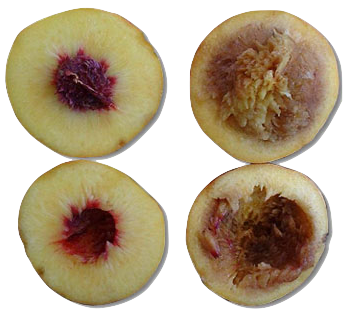


 Chilean wine travels long distances to final markets
Chilean wine travels long distances to final markets
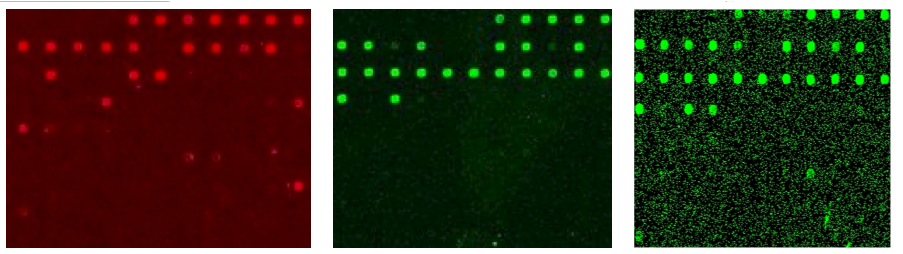
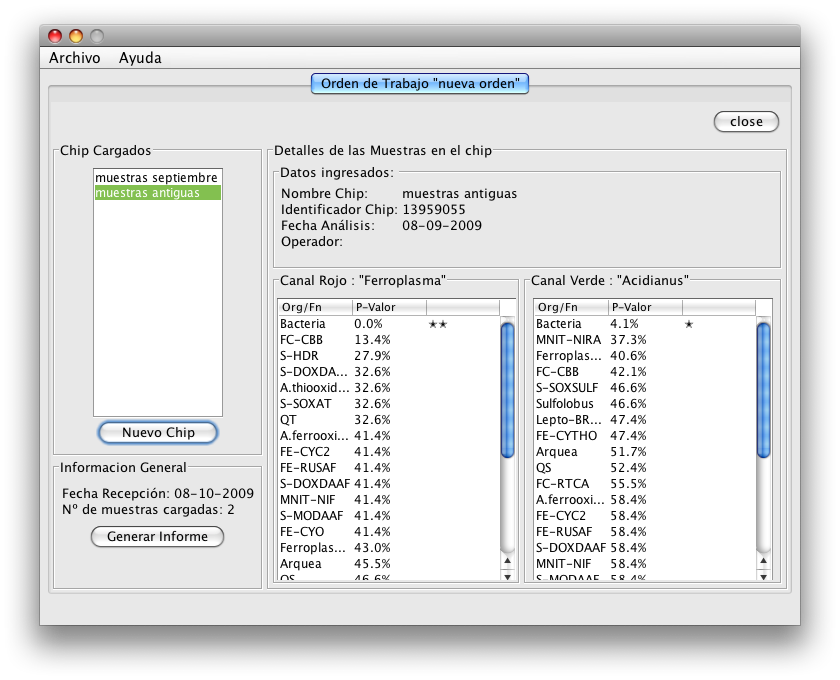
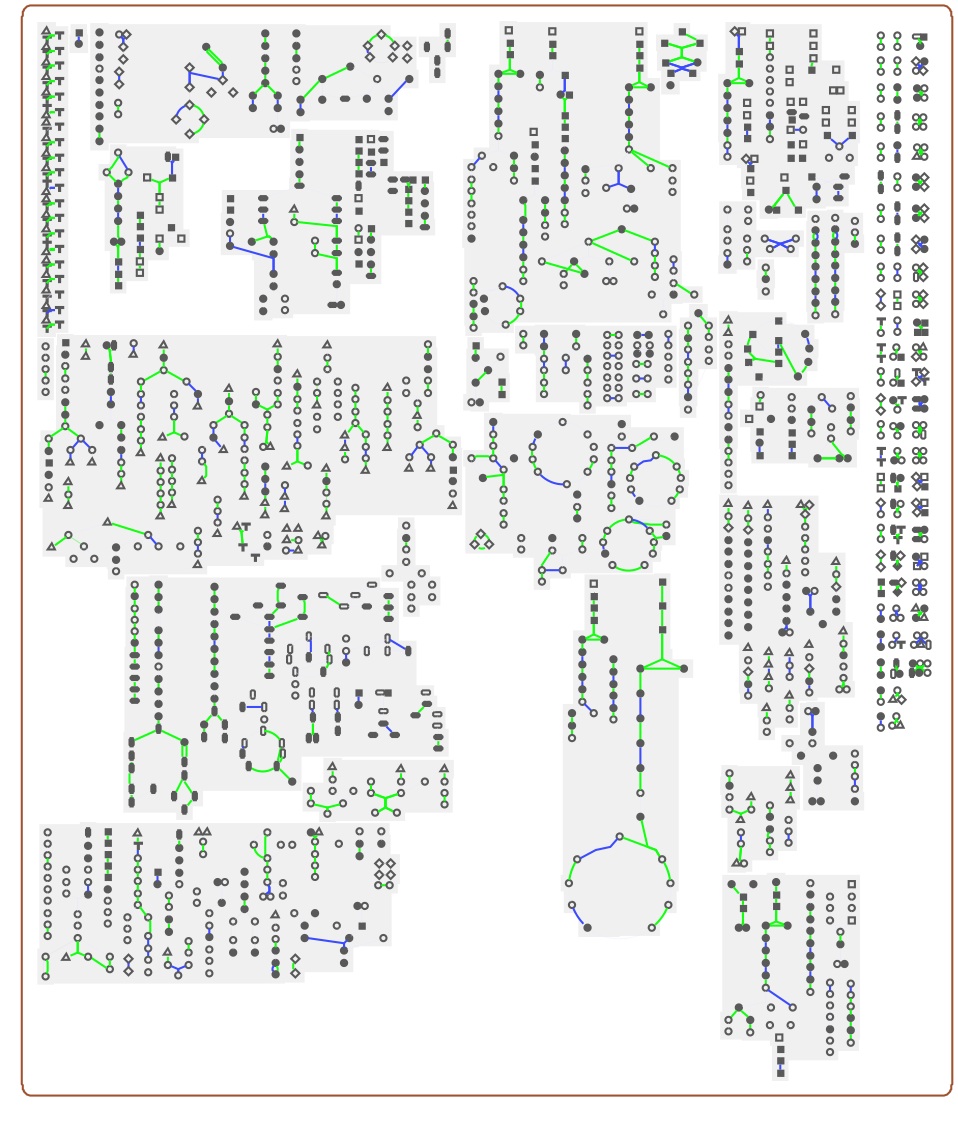 We predict which genes code enzymes
We predict which genes code enzymes