Welcome back
to “Computing for Molecular Biology 1”
We learned how todo this
 How did we do that?
How did we do that?
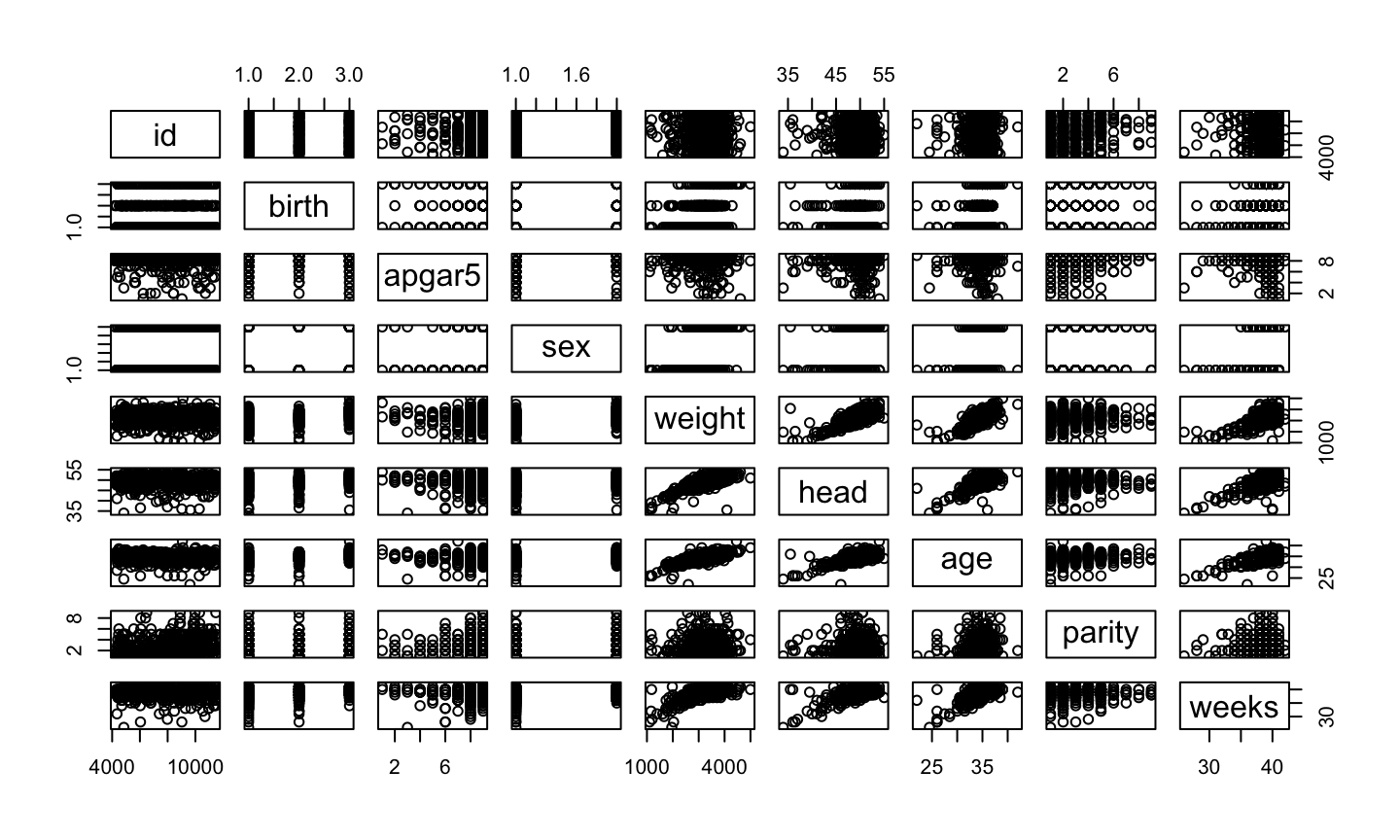
Exploring all data
plot(birth)
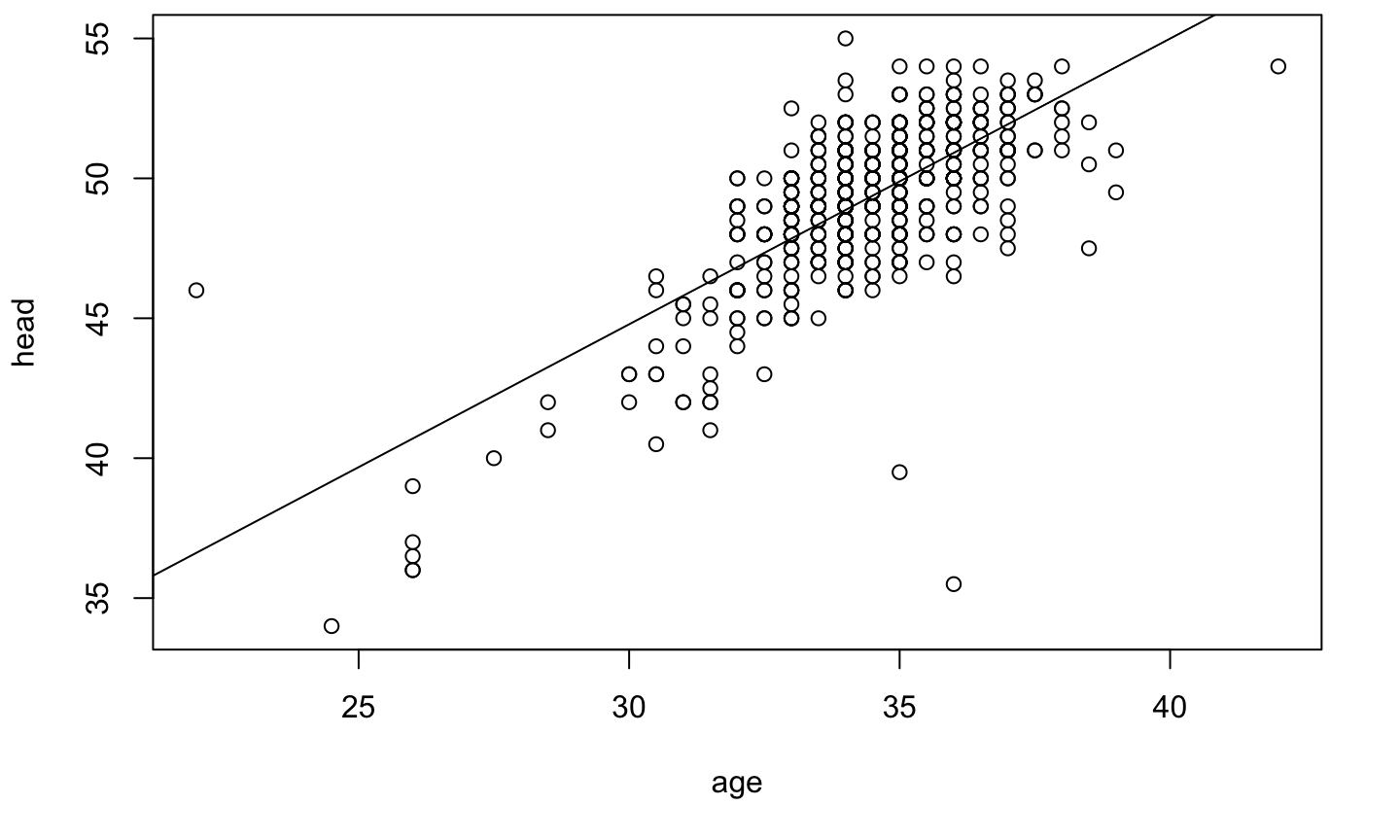
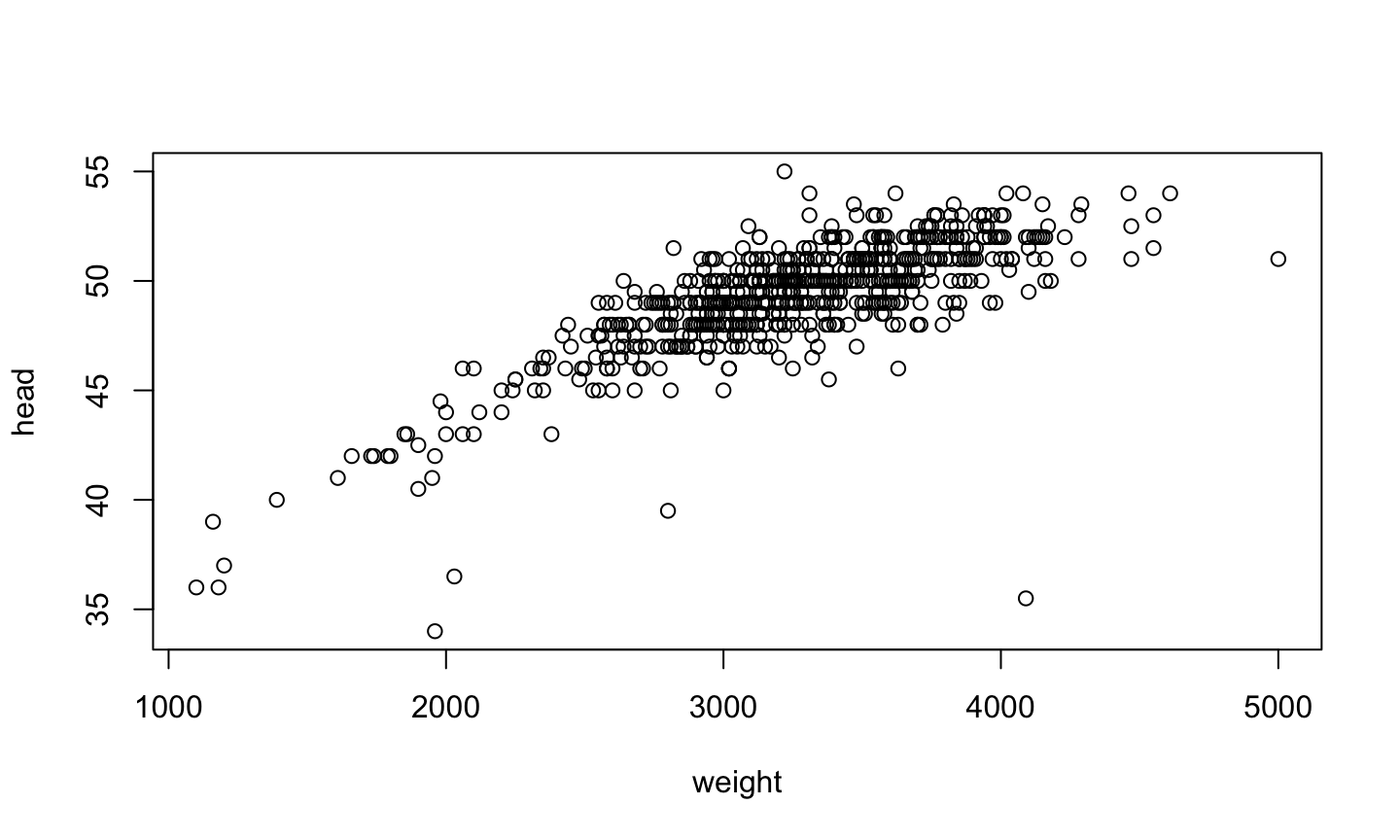
Numeric v/s Numeric
plot(head ~ weight, data=birth)
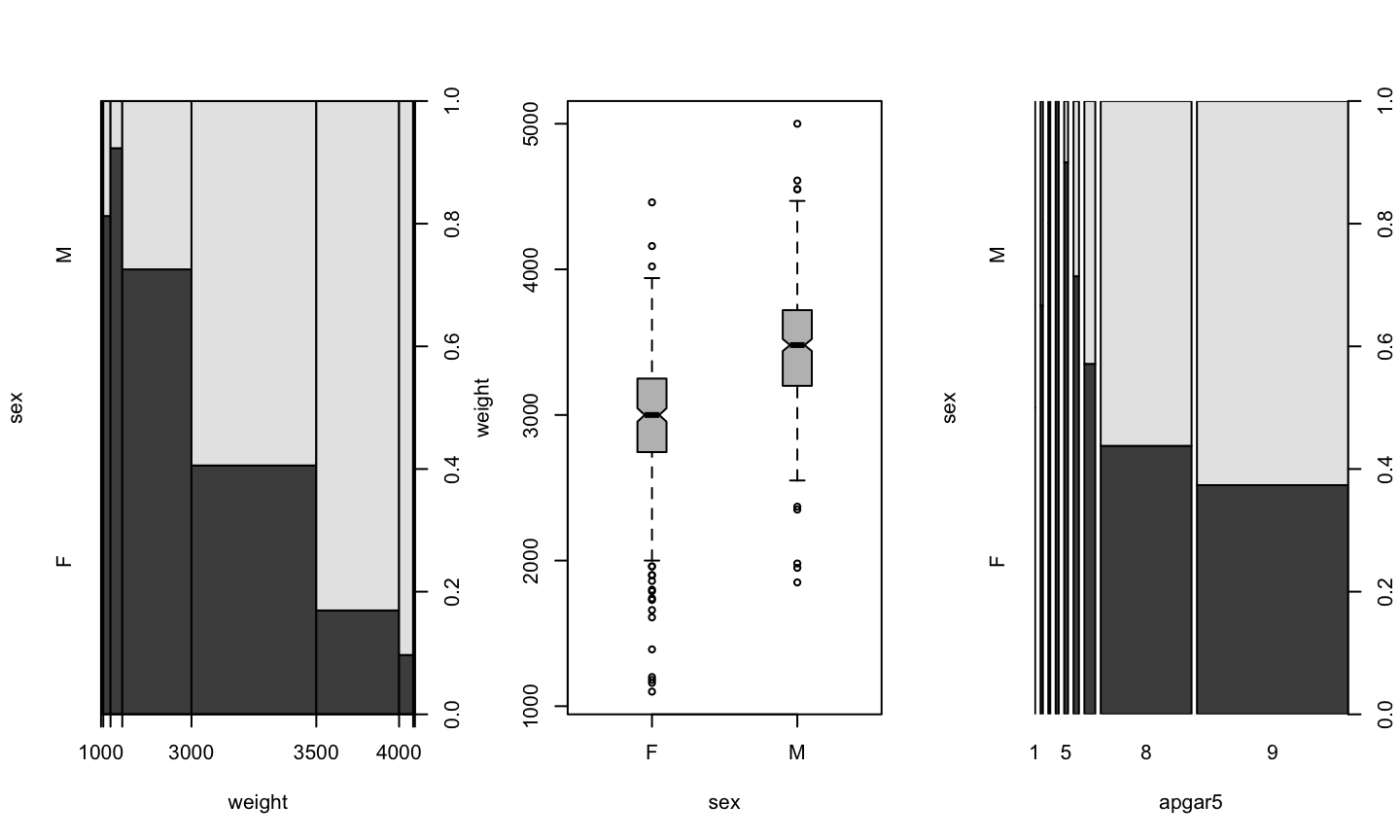
Numeric v/s Factor
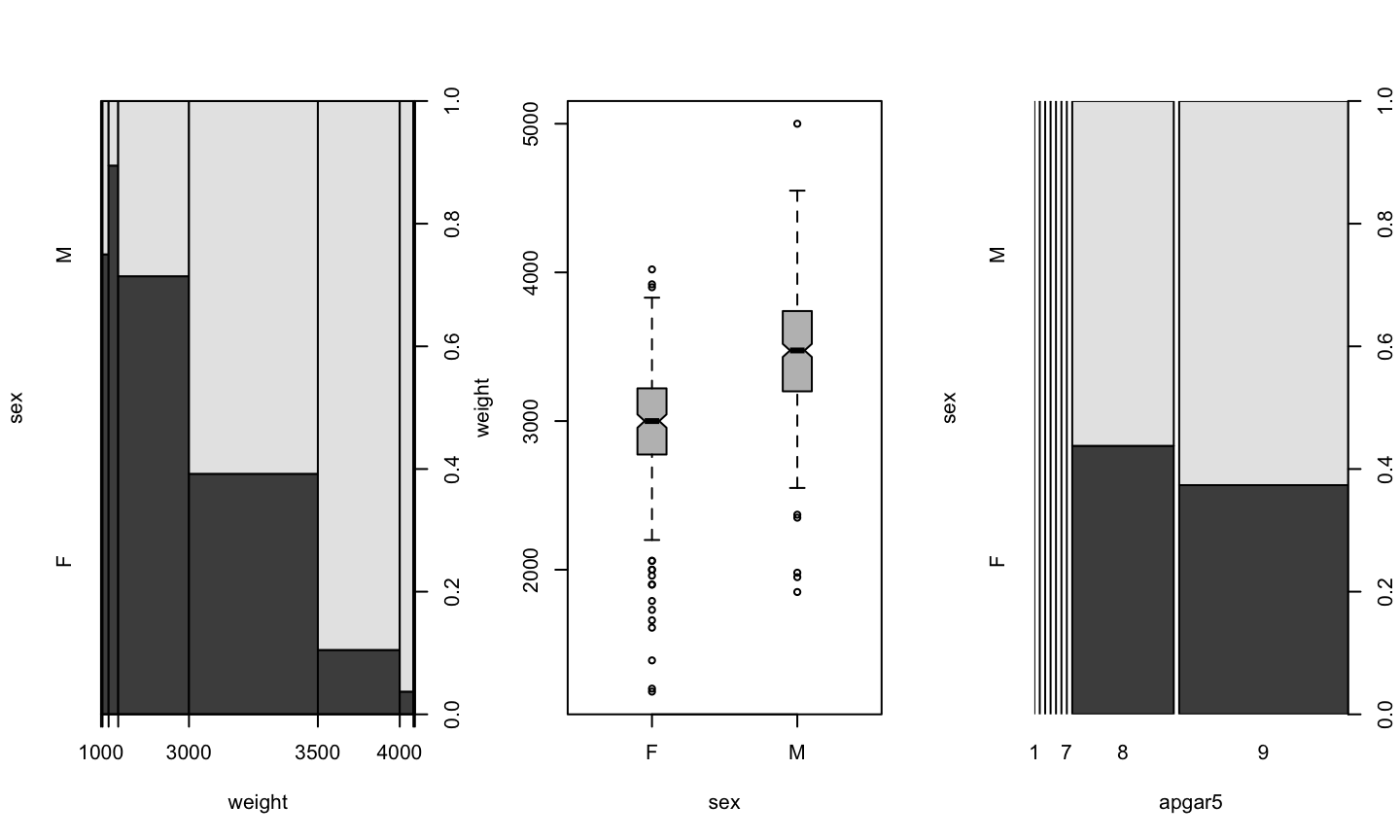
plot(weight ~ sex, data=birth)
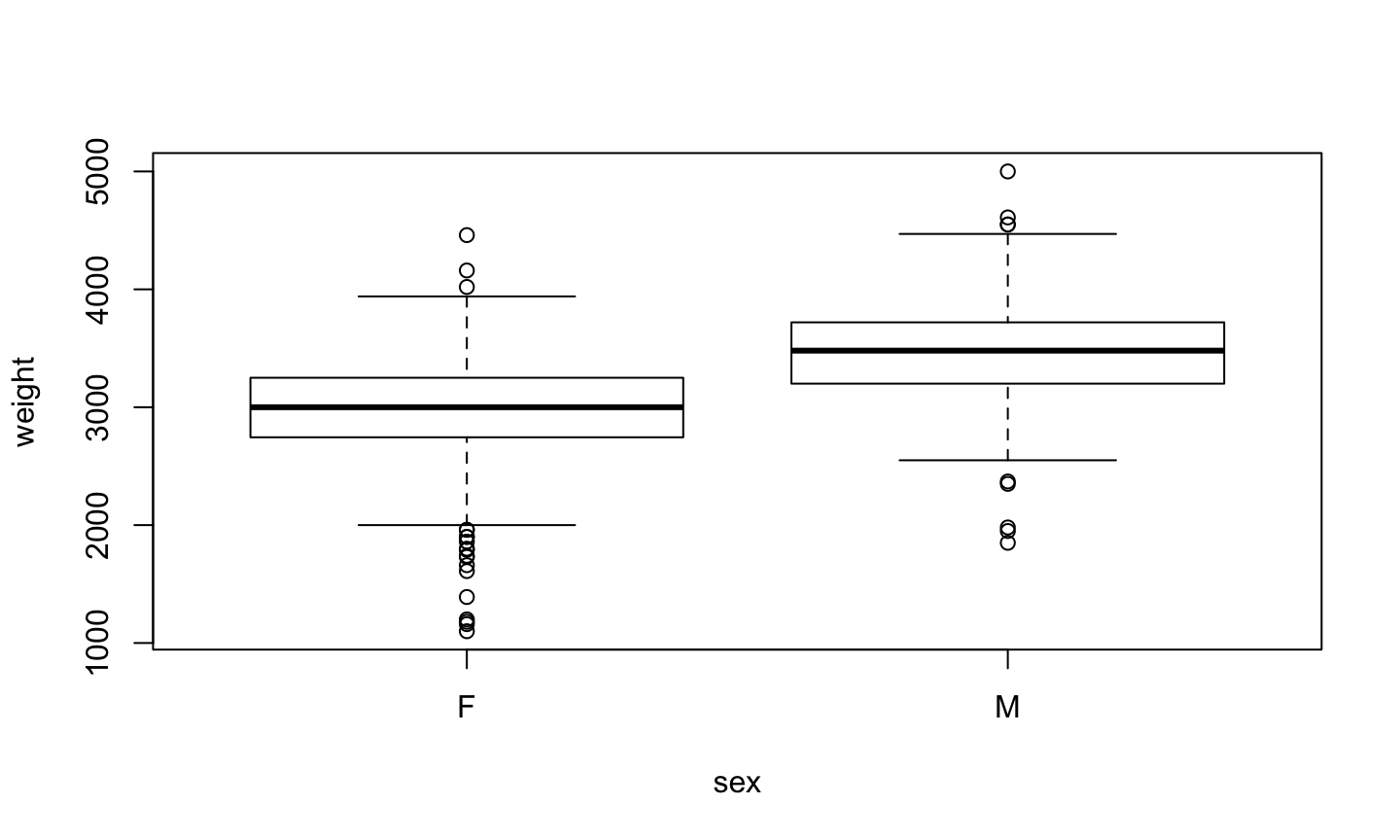
Boxplot
Plotting a numeric value depending on a factor results in a boxplot
It is a graphical version of summary().
- The center is the median
- The box is between the first and third quartil (50% of cases)
- The wiskers extend a prediction of 95% of cases
- Points are outliers
Nicer boxplot
plot(weight ~ sex, data=birth, boxwex=0.2, notch=TRUE, col="grey")
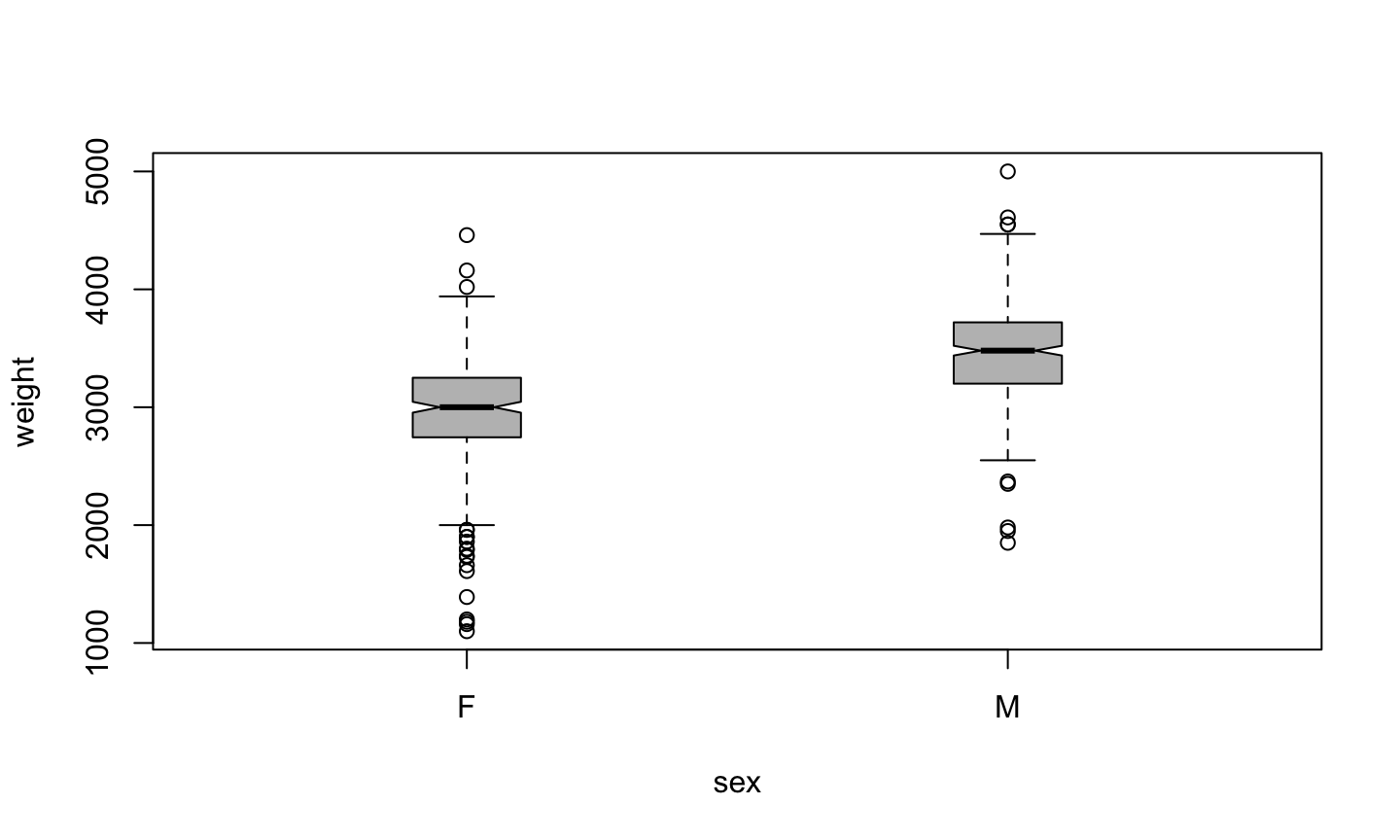
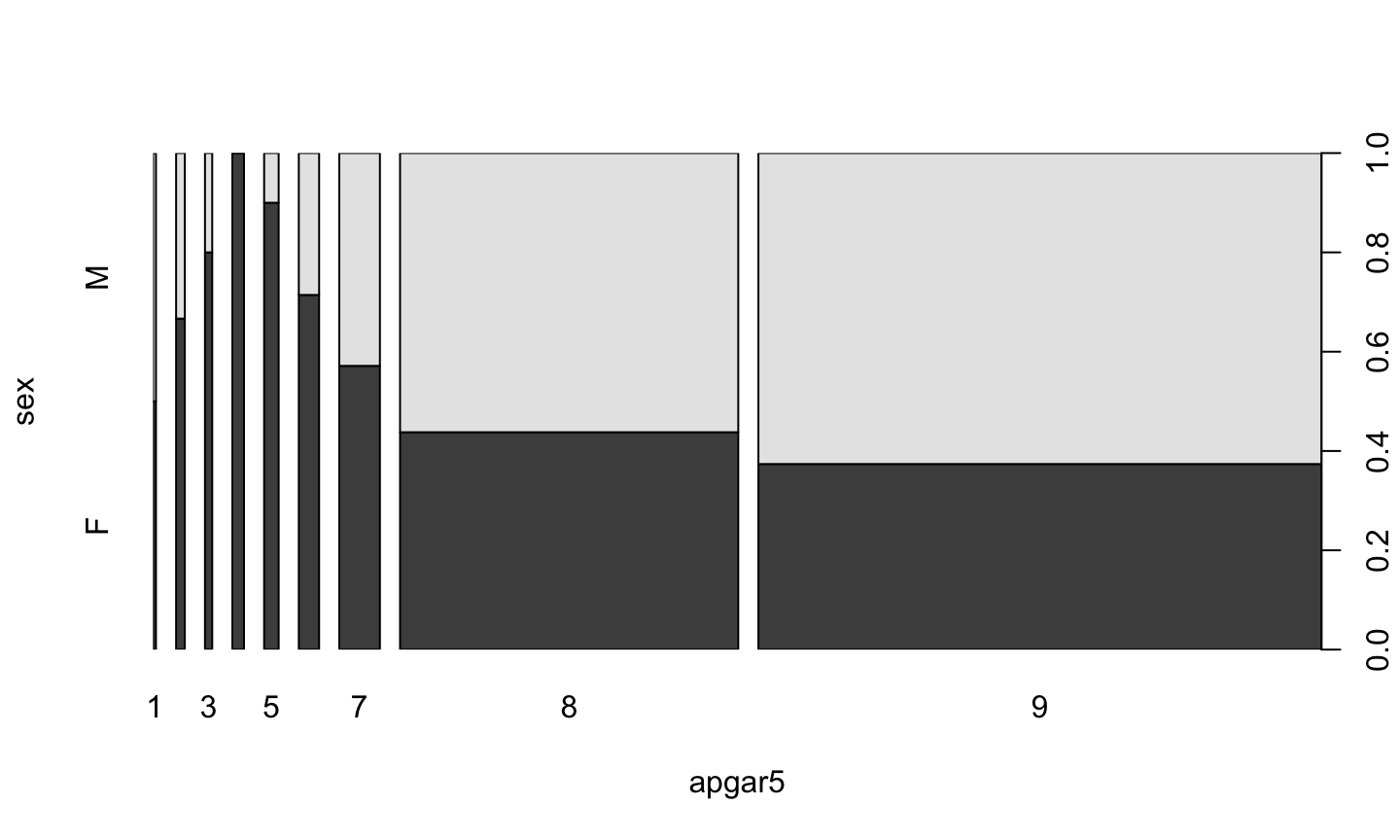
Factor v/s Factor
birth$apgar5 <- as.factor(birth$apgar5) plot(sex ~ apgar5, data=birth)
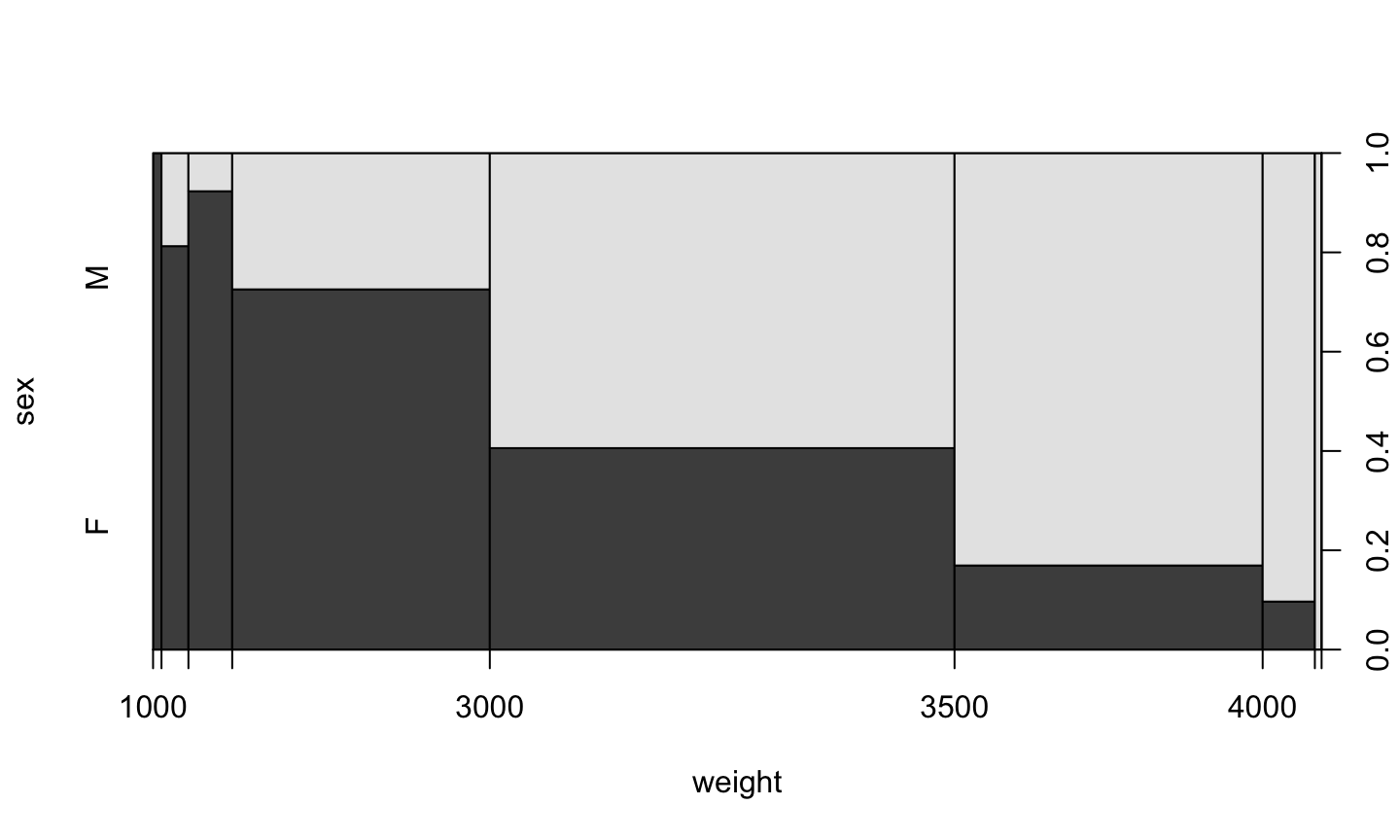
Factor v/s Numeric
plot(sex ~ weight, data=birth)
Saving graphics
Graphical Devices
By default all plot() commands work on a RStudio window
We can open a new device and redirect plot() output
Try this and check the Files window
pdf() plot(sex ~ weight, data=birth) dev.off()
Don’t forget to close the device!!
PDF, PNG, OMG!
There are many outpit devices
The most used are pdf() and png()
Try this and check the Files window
png() plot(sex ~ weight, data=birth) dev.off()
Don’t forget to close the device!!
What is the difference?
- PNG is a bitmap format: a bidimensional array of pixels
- other examples are JPG, TIFF, GIF
- PDF is a vectorial format: a mathematical description
- other example is SVG
The difference is seen when you zoom in
- PDF is good to print in paper
- PNG is better for screen and presentations
Multiple plots
Try this:
pdf() plot(sex ~ weight, data=birth) plot(weight ~ sex, data=birth, boxwex=0.2, notch=TRUE, col="grey") plot(sex ~ apgar5, data=birth) dev.off()
and look at the files.
What do you see?
Multiple plots
Now try this:
pdf(onefile=FALSE) plot(sex ~ weight, data=birth) plot(weight ~ sex, data=birth, boxwex=0.2, notch=TRUE, col="grey") plot(sex ~ apgar5, data=birth) dev.off()
and look at the files.
What do you see?
PDF options
pdf(file = ifelse(onefile, "Rplots.pdf", "Rplot%03d.pdf"),
width, height, onefile, family, title, fonts, version,
paper, encoding, bg, fg, pointsize, pagecentre, colormodel,
useDingbats, useKerning, fillOddEven, compress)
- paper:
- “a4”, “letter”, “legal”, “executive”, “special”, “default”. Defaults to “special”
- width, height:
- specified in inches. Used when paper is “special”
- file:
- filename of the output file. Will be overwritten
PNG options
png(filename = "Rplot%03d.png",
width = 480, height = 480, units = "px", pointsize = 12,
bg = "white", res = NA, ..., type, antialias)
- width, height:
- figure size, in “units”
- units:
- Can be px (pixels, the default), in (inches), cm or mm.
- res:
- The nominal resolution, in pixels per inch (ppi). Default 72.
- pointsize:
- the default pointsize of plotted text
- bg:
- the initial background colour: can be “transparent”.
But ploting many times is boring…
Functions in R
Whenever we need to execute the same set of commands more than 2 times, it can be useful to define a function
The format is:
new.function <- function(options) {
command
command
....
command
return(value)
}
Example
three.plots <- function() {
plot(sex ~ weight, data=birth)
plot(weight ~ sex, data=birth, boxwex=0.2, notch=TRUE, col="grey")
plot(sex ~ apgar5, data=birth)
}
Using it
Try this, line by line:
three.plots()
pdf(file="three-plots.pdf", onefile=TRUE) three.plots() dev.off()
pdf(file="three-plots.pdf", onefile=TRUE) par(mfrow=c(3,1)) three.plots() dev.off()
Using it again
Try this now, line by line:
par(mfrow=c(3,1)) three.plots()
png() three.plots() dev.off()
png() par(mfrow=c(3,1)) three.plots() dev.off()
Passing values to the function
What if we want the same plots for different data?
For example, let’s define
healthy <- subset(birth, apgar5=="8" | apgar5=="9")
(how else can we build the data frame healthy?)
How can we draw the same plots for this data?
New data
summary(healthy)
id birth apgar5 sex weight
Min. : 4199 Min. :1.000 9 :388 F:247 Min. :1180
1st Qu.: 6023 1st Qu.:1.000 8 :233 M:374 1st Qu.:2980
Median : 7894 Median :1.000 1 : 0 Median :3250
Mean : 7836 Mean :1.667 2 : 0 Mean :3255
3rd Qu.: 9601 3rd Qu.:2.000 3 : 0 3rd Qu.:3570
Max. :11475 Max. :3.000 4 : 0 Max. :5000
(Other): 0
head age parity weeks
Min. :35.50 Min. :22.00 Min. :1.000 Min. :29.00
1st Qu.:48.00 1st Qu.:33.50 1st Qu.:1.000 1st Qu.:38.00
Median :49.50 Median :34.50 Median :2.000 Median :39.00
Mean :49.37 Mean :34.45 Mean :2.599 Mean :38.81
3rd Qu.:51.00 3rd Qu.:35.50 3rd Qu.:4.000 3rd Qu.:40.00
Max. :55.00 Max. :39.00 Max. :9.000 Max. :42.00
Redefining three.plots()
three.plots <- function(input) {
plot(sex ~ weight, data=input)
plot(weight ~ sex, data=input, boxwex=0.2, notch=TRUE, col="grey")
plot(sex ~ apgar5, data=input)
}
New function
par(mfrow=c(1,3)) three.plots(healthy)
But now…
three.plots()
Error in eval(m$data, eframe): argument "input" is missing, with no default
It doesn’t work as before
Redefining three.plots() again
three.plots <- function(input=birth) {
plot(sex ~ weight, data=input)
plot(weight ~ sex, data=input, boxwex=0.2, notch=TRUE, col="grey")
plot(sex ~ apgar5, data=input)
}
And now…
par(mfrow=c(1,3)) three.plots()