The Challenges of Modern Molecular Biology and Genetics
The Department of Molecular Biology and Genetics at Istanbul University was created 13 years ago. In the following years it established itself as a well respected department. It attracts every year between 40 and 60 of the best students in the country. They can later follow Masters and Doctorate post-degrees. It has published 376 papers, mainly on plant genomics and fission yeast as a model for human metabolism. Our department is young but is up-to-date with modern molecular biology and genetics theory and practice.
During the same years, we have seen an increasing interest of the general public in molecular biology and genetics. The sequencing of the human genome, made public by the president of USA, captured the attention of everybody. Physicists, mathematicians, computer scientist and engineers, turned their attention to molecular biology questions. They come looking with new eyes and creating new theoretical and practical tools.
http://mediacenter.23andme.com/en-gb/investors/
http://www.ibm.com/podcasts/howitworks/091806/HIW_09182006.pdf
Molecular Biology and Genetics have become mainstream
Today genomic tools are also used outside academia. Several companies provide “personalized DNA services”. They sell genome analysis kits to report hereditary health conditions and traits. One of these companies, 23andMe, is partially owned by Google.
Another case is the Genographic project, created by the National Geographic Society and IBM. It aim to trace ancestry and migrations of the human population. Any person can know which are his true origins.
In these and other cases companies outside academia use molecular biology and genetics tools for their business.
Molecular Biology and Genetics are on the market, and people are joining in
Internet makes Molecular Biology and Genetics accessible to more people
Before Internet times, top science was accessible only to researchers with money to make complex experiments or to buy books and journals. Finding references used to take several weeks by regular mail. Professors had the only copy of the textbooks.
Today all journals are accessible on-line. We can now download references in minutes at low cost, or free when the article is provided in Open Access mode. Most of the important journals follow this policy. Moreover, the experimental results of each article are also available for free. This data has a structure that makes easy for any trained scientist to process it and find new knowledge.
It is easy today to download structured data from all articles related to a subject and combine them to produce new knowledge. Many of the programs used for this meta-analysis are also accessible in Open Source mode. Anyone can download these programs without cost. Scientist can change the program internal code to solve the specific scientific question.
If the analysis requires big computational power it is possible to have it at low cost by using cloud computers. Companies like Amazon.com and Google sell their spare computer power at low prices. This enables researchers to carry computations that would be unfeasible otherwise.
This democratization of knowledge, known as the flat world [@friedman2007world], provides us with an exciting challenge. We have the opportunity to become a player in the big leagues, in a fair game on a leveled surface. But the same opportunity presents to everyone else.
Rich countries have no longer the monopoly of knowledge. We can read the same books and the same articles, use the same machines and the same programs. Anyone could make the new scientific breakthrough, either in New York, New Delhi or Istanbul.
More players come to the game
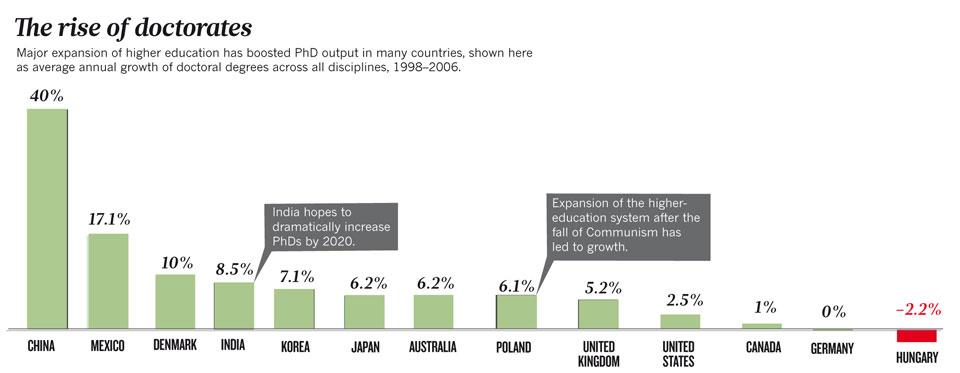
http://www.nature.com/news/2011/110420/pdf/472276a.pdf
The rising of BRICS and MINT economies is also helping to increase the number of researchers worldwide. Today there are more PhD students than ever [@cyranoski2011education]. And many of them will focus on Molecular Biology, Genetics and related areas.
Everyday we see new interdisciplinary collaborations. With other areas of biology, as well as with mathematicians, chemists and computer scientists. It is usual that engineers learn about molecular biology, specially biotechnology engineers.
India graduates more than a million engineers each year. Egypt has 35.000 PhD students and Israel 10.000 [@hays2011phds]. Many of them will find jobs in Molecular Biology companies or academia
Data is cheap, Knowledge is expensive
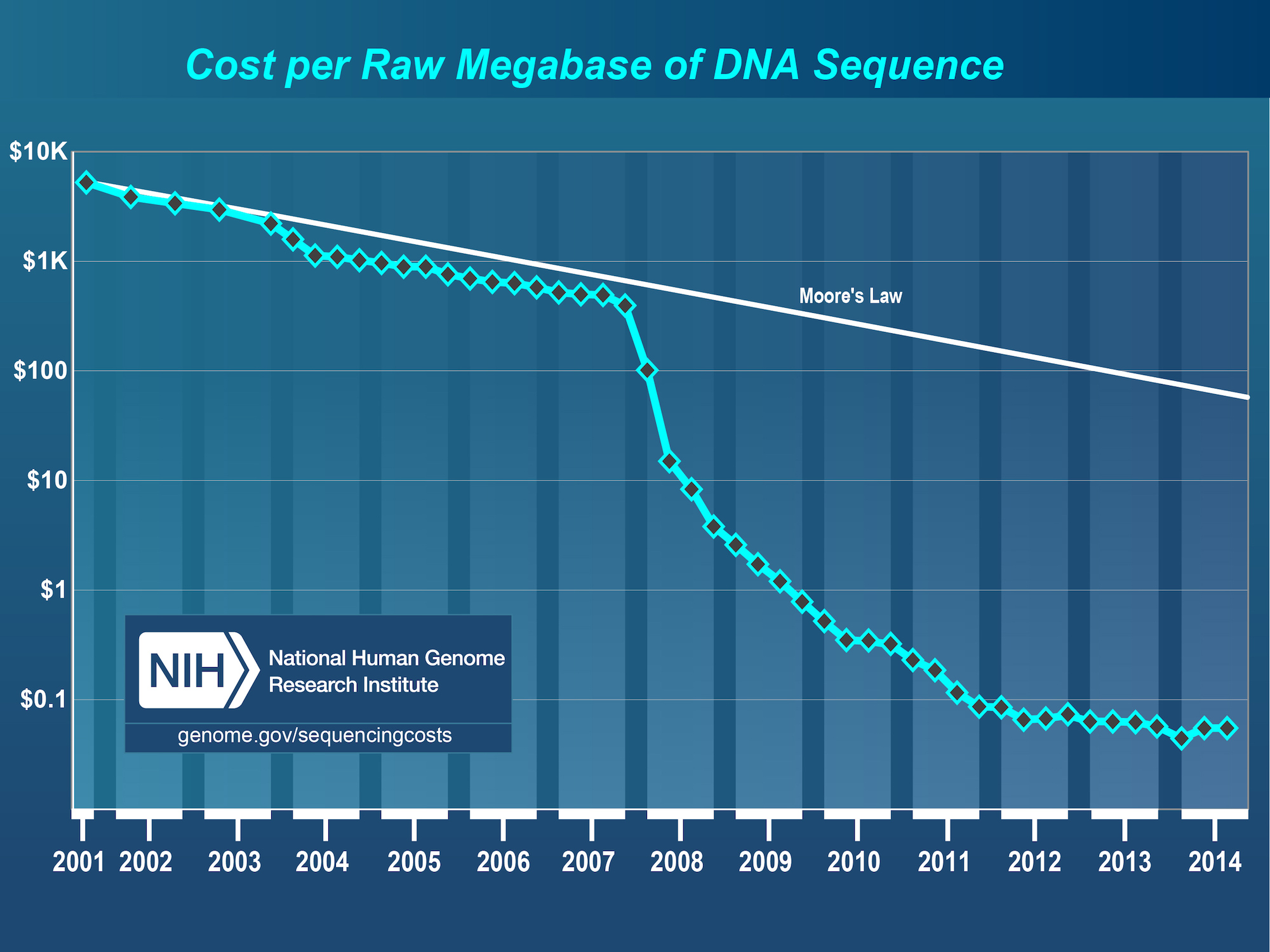
The data production costs are reducing everyday. The value is on the ideas. Advances in molecular biology and genetics have resulted in a new generation of efficient machines. Instruments such as new generation sequencers, microarrays, mass spectrometers and real-time PCR. They produce reproducible experimental results in big volumes and at low cost.
A well planned experiment can be outsourced to an external lab. The value is on the plan, the selection and handling of the samples and in the analysis. Data without analysis will not have any impact.
Research changes from producing Data to producing Information, Knowledge and Wisdom
PCR can soon become available for the general public

Today PCR thermocyclers are expensive devices found in universities and research centers, very much like desktop computers were in the 70’s and 80’s. Nowadays computers are low-cost and found everywhere.
Will the same happen with PCR?
PCR thermocyclers can be really low-cost. There is no technical reasons why they cannot be made as low-cost as an Arçelik Mini Telve Turkish coffee maker. They have essentially the same pieces.
Today only a few companies produce PCR thermocyclers, just like smartphones such as the iPhone and Samsung. Nevertheless you can see them everywhere. And this is a big opportunity for creators of software applications.
PCR is the new PC
“A computer on every desk and in every home, all running Microsoft software”. Bill Gates, Microsoft’s founding mission.
Gates set this goal in the late 70’s, when it was not obvious if people would even see a computer in their lives [@stross1997microsoft;@fortune1995]. PCR technology is now in the same state that Personal Computers were in 1975.
If PCR machines become inexpensive, and there is “a PCR on every desk and home”, in hospitals, restaurants and high schools, then who will be making “software apps” for them?
What will be our competitive advantage in the next ten years?
If we don’t fully analyze our own experimental data, someone else will do. And they will publish it.
A Proposal to Get Ready for the Next Decade
Good science is the combined result of asking the good questions, selecting and preparing the good samples, carrying on correct experiments, measure the resulting values with the proper instruments, making the most complete analysis, and determine the relevant biological conclusions.
If we keep the current good job, we will perform high quality experiments and produce results in increasing volumes. But data without analysis is becoming less valuable. For example, the first bacterial genomic sequence was published in Science journal [@Fleischmann28071995]; today it would be just a shot communication.
Young molecular biologists need to learn how to extract relevant knowledge and biological meaning from the raw data.
Given the large volume of data and the global competition with other researchers, this can only be done using computers and informatics tools. We should increase the competences of our undergraduate students on data science. They should be at ease using the tools that exist today.
Our graduate level students shall go beyond generic data science tools. They shall integrate molecular biology and analytical methods. Get familiar with “Systems Biology”, the computational and mathematical modeling of complex biological systems. They need to understand how properties emerge in regulatory, metabolic and cell signaling networks. The mathematical tools can be learned together with the biological context, so they make sense and are easier to learn.
Every normal student is capable of good mathematical reasoning if attention is directed to activities of his interest [@piaget1976child]
How will we be different from the rest?
If we use the same instruments, theories and tools as everyone else, we will be just one more lab.
The second way we can use to increase the impact of our science is to design and use new instruments. New tools that can measure faster, with more precision or at lower cost. Many of the most important advances in science are consequence of the creation of new instruments.
At first glance it seems that instruments for molecular biology are high-technology tools that we cannot make or modify. A “space age” technology out of our reach. But all instruments are based on the same basic physicochemical principles. We already know and understand these principles. And current technology allows us to make and modify our own instruments. MIT has being teaching this in the course “How to Make Almost Anything” since 1998. We can also teach it.
We should teach to our students how to make laboratory instruments, and encourage them to create the tools that do not yet exist.
Let us use computers and Internet to quickly learn the current tools
We can and should use all online tools in our advantage. Use online material to teach and learn faster. Students are already doing so.
We shall overcome the language barrier. Science language is English, for good or bad. Young people learn languages fast.
We shall provide tools to enable interdisciplinary collaboration, in particular with data scientists, learn to work online, on distributed or international workgroups, learn about international funding tools and understand intellectual property rules, patents and licenses.
And we should be making our own online teaching material.
Data Analysis for Molecular Biology
- move from creating data to creating information, knowledge and wisdom
- undergraduate level:
- use more online resources
- English writing class
- introduction to data science
- programming for research
- graduate level:
- advanced computational tools
- biostatistics and data mining
- systems biology
- intellectual property
- create a bioinformatics and systems biology laboratory
Undergrad Level Computation Courses
Teaching Data Science
We can transform the “Computation III” course, which today is “Databases”, into an “Introduction to Data Science”.
The students will learn how to handle experimental data and how to communicate with scientists of other data-oriented disciplines.
They will learn how to produce publication quality reports with reproducible results. How to get raw data, extracting relevant information, filter it using several selection criteria. How to store and retrieve it in efficient and useful ways. How to transform it, organize it, categorize it, display, show and understand the results.
Tools include Unix command line tools, SQL and the R statistical package. The student should be able to understand how computer networks work and what are their limitations.
Teaching modern programming
Transform “Computation IV” from “Making Web pages” to “Programming for Research”.
Teach Python and BioPython to analyze, model, evaluate and predict the behavior of genomic and molecular biology entities.
The students should be able to interact with high end servers, use command line tools and be comfortable in computing environments others than Microsoft Windows.
The objective of this course is no to make our students experts on computer science, but to give them the concepts and language that will allow them to collaborate in interdisciplinary groups
Graduate Level Courses
Preparing our people for the molecular biology to come
Communication
If current global trends continue, the new generations of molecular biologist will work in frequent collaboration with peers from other countries and other disciplines. They need to have not only a good level of English, but they also need to handle the language and concepts of other disciplines. We can help them by creating “Scientific communication workshops” where they can exercise and improve their writing and speaking skills. In this workshop students should practice writing papers and present them, in English, as a rehearsal of a conference. It can be shared with other departments to encourage multidisciplinary interaction and learning of technical language.
Collaboration also requires online tools. Papers will be written by many authors simultaneously using Dropbox, Google Docs or GitHub. Or maybe it will be more like Wikipedia or arXiv. Our students need to know these tools and the methodology that each one requires.
Intellectual property
Many of these collaborations and publications will not be necessarily published in scientific journals. Some may be patented, some may be licensed for industrial applications. Others may be published on Wikipedia or other web media. Or delivered as open source. The alternatives are many and they are relevant for the new generation of molecular biologists. There should be a lecture where the students learn what can be patented, how to patent an invention and about the different licenses that protect or disclose intellectual property.
Systems Biology and Advanced Computational Tools
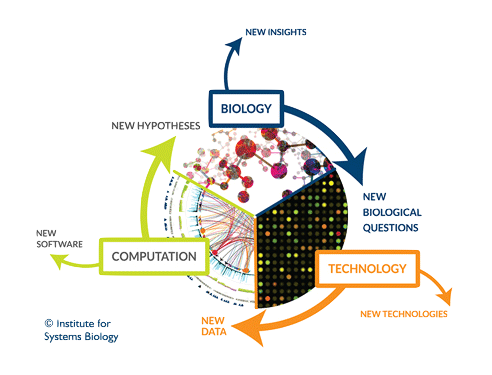
Analysis of Complex Biological Systems
As long as molecular biology and genetics experiments continue to produce big volumes of results, the scientific value shifts from the data to the ideas. If a researcher wants to show that apples are good for health, it is not longer enough to show statistical data that correlates apple consumption and health indices. He need to propose a model of the way that apples improve health, design an experiment to prove these hypothesis versus alternative ones and analyze that experiment.
One approach that appeared in the last years for doing this on molecular biology is “Systems Biology”, the study of the interactions between the components of biological systems, and how these interactions give rise to the function and behavior of that system. This approach combines biology theory with mathematical and computational models.
Our students need to know the basic concepts of statistical analysis and mathematical modeling used in Systems Biology, and computational concepts for carry out these models by themselves or in collaboration.
Advanced Computing Concepts
Grad students that follow a specialization on Systems Biology or Computational Biology will be interacting with scientists of other areas. They need the language and concepts of parallel and distributed computing, clusters, cloud, massive parallel computation, artificial intelligence, neural networks, genetic algorithms and other topics of high performance computing and data mining. We should provide the environment where they can grasp these concepts. Either an “Advanced Computing” lecture, a workshop or a laboratory.
Creating a “Laboratory of Bioinformatics and Systems Biology”
To consolidate the lessons given in the previously discussed courses, we need a physical space where the students can put them in practice. This would be a laboratory like the others already existing in our department, only with different kinds of tools.
In a first stage it would be ideal to have
- Books and bookshelves
- Physical space, network connection and electrical power for near 6 people, their computers and other devices
- Wifi access point
- Color laser printer
- Desktop computers (Linux and Apple)
- Linux server
- Whiteboard
- Data projector or flat TV
- Table
- Chairs
We will not teach math “just-in-case” it is useful. Students will learn “just-in-time” when they need it. The idea is the teaching be driven by the demand of knowledge, not by the supply of it.
Theory of constructivism suggests that learners construct knowledge out of their experiences. Learners with different skills and backgrounds should collaborate in tasks and discussions to arrive at a shared understanding of the truth in a specific field [@duffy1992constructivism].
New tools for new science
Creating innovative experimental tools

Main advances in Molecular Biology and Genetics (and in other sciences) are often the result of using new instruments, which usually are named according to their inventor
- Galileo created modern science when he made his own telescope
- Newton also invented a new kind of telescope, still used today
- Bunsen enabled spectrometry analysis with his burner
- Svedberg ultracentrifugue
- Sanger DNA sequencing method
- Southern blot method for specific DNA detection
- PCR to amplify DNA samples
Notice that most of these inventors got Nobel prizes for their contributions.
Usually we think that building molecular biology tools is beyond our capabilities. Nevertheless in the last 5 years this has changed dramatically.
Today low-cost 3D printers, laser-cutters and computer controlled manufacturing tools are used by amateurs scientist and technology enthusiast around the world to make all kind of advanced and complex devices [@anderson2012makers].
Here in Istanbul we find several low-cost laser-cutter companies, 3D printers are sold in shopping malls (as seen in the picture) and there are at least two “makerspaces”, social clubs where anyone can teach, learn and use these technologies.
Using the same instruments on the same organisms as others is like being a cover band. We can play well but we will not make a real impact
Making “Software” for PCR machines

We have seen, on computers and smartphone industries, that once the hardware becomes widely available, the software industry flourishes. As we discussed earlier, PCR are now ready to follow the same path.
If PCR machines are available everywhere, applications can be:
- Determining ancestry (e.g. race horses, farm animals, fishes)
- Detection of unwanted organisms
- Marker-assisted breeding
- Targeted expression analysis
- Food quality control in restaurants (for example in an university canteen)
- Security and control of Genetically Modified Organisms
- Polymorphism detection
- Clinical diagnosis
- Personalized medicine
- Police forensic analysis
- Choosing strains that maximize yield in biotechnology industry
Making the new “apps” makers
Software for PCR means the specific parameters of an application:
- DNA extraction protocols
- Primers design
- Amplification protocols
- Detection methods
We should prepare our students to make these “apps”. They should have easy access to low-cost thermocyclers, use them frequently and creatively.
Then, like in the computer industry, they may create completely new applications that we cannot foresee now.
We can make our own low-cost PCR machines based on the OpenPCR model. The design is free, materials are inexpensive.
Learning by Making
Amateur scientists are already building low cost PCR machines. For example OpenPCR is a kit with cost under USD$600. Its design files can be downloaded from the web and built at home.
If PCR machines are open source now, which tool will be the next?
We can create an advanced course on “Scientific Instrumentation” using initially software tools. In some subjects we can partner with the Electrical Engineering Department.
It is easy to enter in this area. There is no need for a huge initial investment. Making instruments is now “software”, not craftsmanship.
When needed we can first use the services of the existing “makerspaces”. If our prototypes are successful, then we can have our own 3D printer or laser cutter.
Online collaboration enables fast change of designs.
Computer aided manufacturing tools transform these designs into real objects. We can understand this with a biological analogy.
Designs in digital files are like genes. 3D printers are like ribosomes, producing physical versions of the design. Online collaboration is like the evolution: designs are changed to improve their fitness.
A “Scientific Instrumentation” course should also consider teaching the use of online collaboration tools.
The best tools result from iterative improvement of pre-existing designs. Just like software. And science.
Creating a “Scientific Instrumentation Laboratory”
Initially the theoretical part of an “Scientific Instrumentation Laboratory” can be developed in the same space of the “Systems Biology Laboratory”. The basic principles can be learned in periodic workshops, where each participant can teach and learn. Prototype designs can be made in the computer. The physical versions of these prototype tools can be fabricated in collaboration with the existing “makerspaces” or outsourcing to laser-cutters and 3D printer services.
In the second year we will evaluate the economical feasibility of having our own 3D printer and other maker tools.
Funding
Most of the ideas presented in this executive summary are zero-cost or low-cost. The only major investment would be this laboratory. The details of the costs are described in an appendix. In summary, besides the physical space, the cost is between USD $10.000 and $20.000. Specific funding sources (national or European) are open to discussion.